6LIH
 
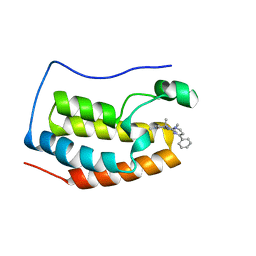 | | BRD4 BD1 bound with compound 10 | | Descriptor: | (3~{R})-1,3-dimethyl-6-[(4-phenylpyrimidin-2-yl)amino]-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62030542 Å) | | Cite: | BRD4 BD1 bound with compound 10
To Be Published
|
|
6L9H
 
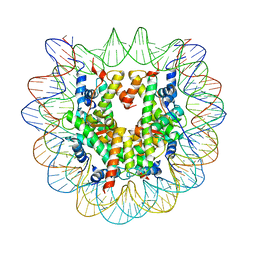 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6LA9
 
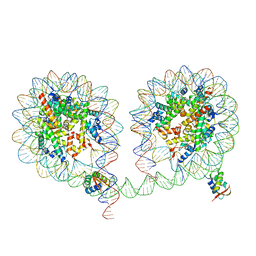 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6L62
 
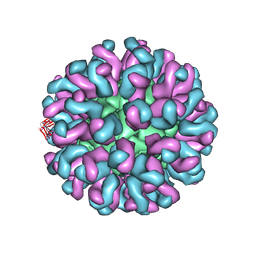 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6L9Z
 
 | | 338 bp di-nucleosome assembled with linker histone H1.X | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LAB
 
 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LER
 
 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6ZLY
 
 | |
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
6LCI
 
 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
6LAG
 
 | | Solution structure of SPA-2 SHD | | Descriptor: | Spa2-like protein | | Authors: | Fan, J.S, Wong, J.Y, Zheng, P, Yang, D, Jedd, G. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spitzenkorper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds.
Nat Commun, 11, 2020
|
|
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
6ZGQ
 
 | | AceL NrdHF class 3 split intein GSH linked splice inactive variant - C124A, N146A | | Descriptor: | AceL NrdHF-1-1 Intein, IODIDE ION | | Authors: | Hoffmann, S, Mootz, H.D, Kuemmel, D, Singh, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of an Unusual and Naturally Split Class 3 Intein.
Chembiochem, 22, 2021
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6LP2
 
 | | Structure of Lpg2148/UBE2N-Ub complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 N, Uncharacterized protein lpg2148 | | Authors: | Feng, Y, Wang, Y, Huang, Y, Li, D. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structure of Lpg2148/UBE2N-Ub complex
To Be Published
|
|
8FXC
 
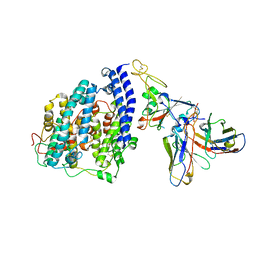 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6M2W
 
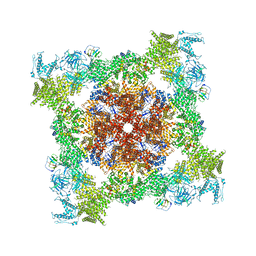 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6LS4
 
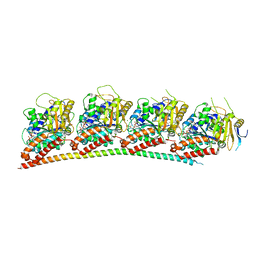 | | A novel anti-tumor agent S-40 in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(4-cyclopropylphenyl)sulfonylamino]-4-methyl-N-(pyridin-3-ylmethyl)benzamide, GLYCEROL, ... | | Authors: | Du, T, Lin, S, Ji, M, Xue, N, Liu, Y, Zhang, K, Lu, D, Chen, X, Xu, H. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity.
Cancer Lett., 495, 2020
|
|
6LT6
 
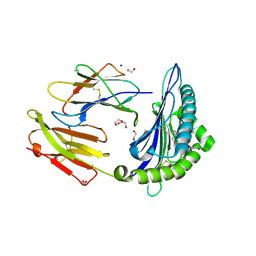 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LZL
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with Piperine | | Descriptor: | (2E,4E)-5-(2H-1,3-benzodioxol-5-yl)-1-(piperidin-1-yl)penta-2,4-dien-1-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Liu, Z.H, Wu, D, Lu, W.Q, Huang, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with Piperine
To Be Published
|
|
6LNU
 
 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
6MBN
 
 | | LptB E163Q in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Owens, T.W, Ruiz, N, Kahne, D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Combining Mutations That Inhibit Two Distinct Steps of the ATP Hydrolysis Cycle Restores Wild-Type Function in the Lipopolysaccharide Transporter and Shows that ATP Binding Triggers Transport.
Mbio, 10, 2019
|
|
7O8K
 
 | | NmHR light state structure at 300 us after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
