1UHV
 
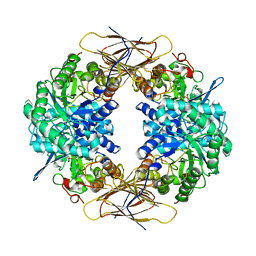 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
2N8N
 
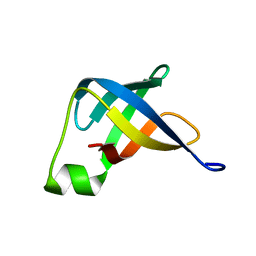 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
1PX8
 
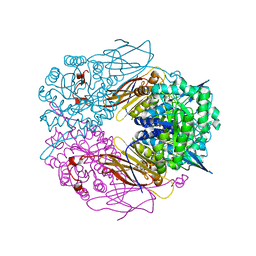 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
6DQ1
 
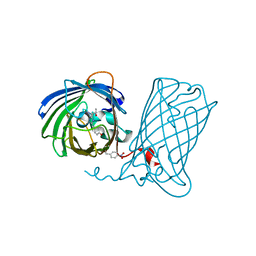 | |
6DQ0
 
 | | sfGFP D133 mutated to 4-nitro-L-phenylalanine | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, superfolder green fluorescent protein | | Authors: | Phillips-Piro, C.M, Maurici, N, Lee, B. | | Deposit date: | 2018-06-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of green fluorescent protein with the unnatural amino acid 4-nitro-L-phenylalanine.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7ZDD
 
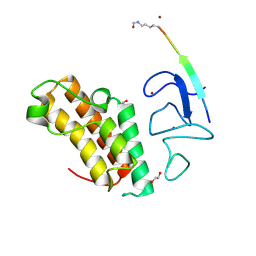 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K10ac histone peptide. | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM33, Histone H3.X, ... | | Authors: | Caria, S, Duclos, S, Crespillo, S, Errey, J, Barker, J.J. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Identification of Histone Peptide Binding Specificity and Small-Molecule Ligands for the TRIM33 alpha and TRIM33 beta Bromodomains.
Acs Chem.Biol., 17, 2022
|
|
7ZM5
 
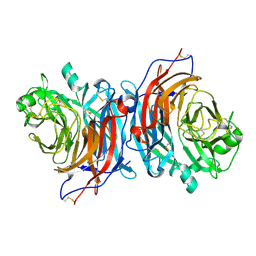 | | Structure of Mossman virus receptor binding protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein | | Authors: | Stelfox, A.J, Bowden, T.A, Rissanen, I, Harlos, K. | | Deposit date: | 2022-04-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution state of the C-terminal head region of the narmovirus receptor binding protein.
Mbio, 14, 2023
|
|
5EVM
 
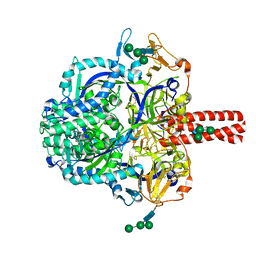 | | Crystal Structure of Nipah Virus Fusion Glycoprotein in the Prefusion State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MALONATE ION, ... | | Authors: | Xu, K, Nikolov, D.B. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.367 Å) | | Cite: | Crystal Structure of the Pre-fusion Nipah Virus Fusion Glycoprotein Reveals a Novel Hexamer-of-Trimers Assembly.
Plos Pathog., 11, 2015
|
|
8RVN
 
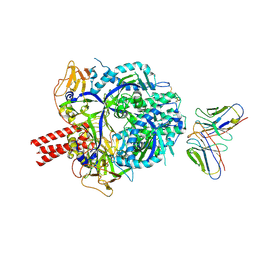 | | Nipah virus (NiV) fusion protein in complex with neutralizing Fab92 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab92 heavy chain, Fab92 light chain, ... | | Authors: | Avanzato, V.A, Stass, R, Duyvesteyn, H.M.E, Bowden, T.A. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A monoclonal antibody targeting the Nipah virus fusion glycoprotein apex imparts protection from disease.
J.Virol., 98, 2024
|
|
1DSF
 
 | |
8EQF
 
 | | cryoEM structure of a broadly neutralizing anti-SARS-CoV-2 antibody STI-9167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Bajic, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | An in vitro experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody.
Mbio, 15, 2024
|
|
2FI7
 
 | |
7RWJ
 
 | |
7RXB
 
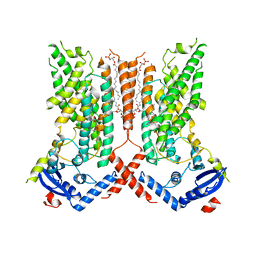 | | afTMEM16 lipid scramblase in C18 lipid nanodiscs in the absence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RX2
 
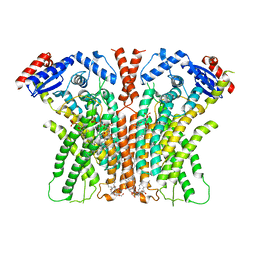 | |
7RXA
 
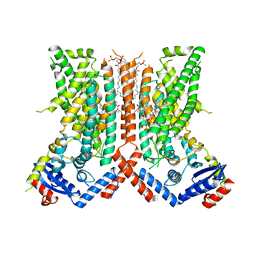 | | afTMEM16 DE/AA mutant in C14 lipid nanodiscs in the presence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RXH
 
 | |
7RX3
 
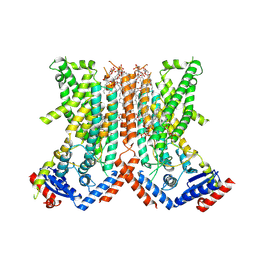 | |
7RXG
 
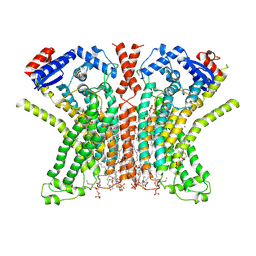 | |
7SKU
 
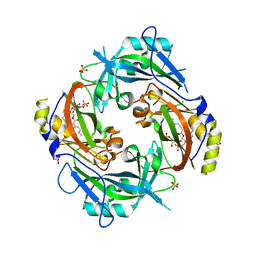 | | Nipah virus matrix protein in complex with PI(4,5)P2 | | Descriptor: | Matrix protein, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Norris, M.J, Saphire, E.O. | | Deposit date: | 2021-10-21 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization.
Sci Adv, 8, 2022
|
|
7SKS
 
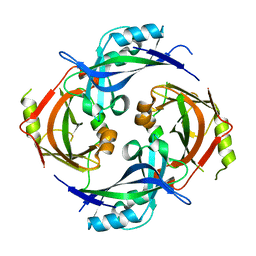 | |
7SKT
 
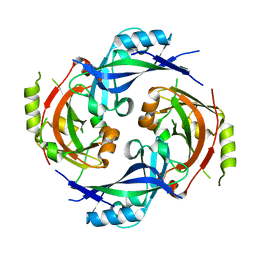 | |
5V4X
 
 | | Human glucokinase in complex with novel pyrazole activator. | | Descriptor: | (2S)-3-cyclohexyl-2-[4-(cyclopentylsulfonyl)-2-oxopyridin-1(2H)-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Skene, R.J, Hosfiled, D.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5V4W
 
 | | Human glucokinase in complex with novel indazole activator. | | Descriptor: | (2S)-2-[4-(cyclopropylsulfonyl)-1H-indazol-1-yl]-N-(5-fluoro-1,3-thiazol-2-yl)-3-(oxan-4-yl)propanamide, Glucokinase, IODIDE ION, ... | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
