1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
1UHR
 
 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
5AZ2
 
 | | Crystal structure of the Fab fragment of 9E5, a murine monoclonal antibody specific for human epiregulin | | Descriptor: | anti-human epiregulin antibody 9E5 Fab heavy chain, anti-human epiregulin antibody 9E5 Fab light chain | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3J3Z
 
 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
4XFP
 
 | | Crystal Structure of Highly Active Mutant of Bacillus sp. TB-90 Urate Oxidase | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Hibi, T, Hayashi, Y, Kawamura, A, Itoh, T. | | Deposit date: | 2014-12-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Glycine Substitution of Surface Proline 287 Involves Entropic Enhancement of Bacillus sp. TB-90 Uricase Activity
To be published
|
|
6ZBO
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | Descriptor: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6ZBN
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in complex with tert-butyl 6-(5-hydroxy-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-1-yl)nicotinate (IOX4) | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6IN0
 
 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN4
 
 | | Crystal structure of apo DAPK1 in the presence of 18-crown-6 | | Descriptor: | Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IMZ
 
 | | Crystal structure of MTH1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN3
 
 | | Crystal structure of DOT1L in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
3IF5
 
 | | Crystal Structure Analysis of Mglu | | Descriptor: | Salt-tolerant glutaminase | | Authors: | Yoshimune, K, Shirakihara, Y. | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product L-glutamate and its activator Tris.
Febs J., 277, 2010
|
|
5B1S
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with 2-(2-fluorophenyl)ethanamine | | Descriptor: | 2-(2-fluorophenyl)ethanamine, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | In silico, in vitro, X-ray crystallography, and integrated strategies for discovering spermidine synthase inhibitors for Chagas disease
Sci Rep, 7, 2017
|
|
3WZP
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZQ
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZO
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | Descriptor: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X00
 
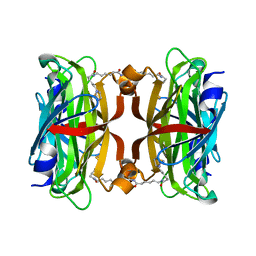 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
8XDT
 
 | |
8Z8O
 
 | |
8ZFW
 
 | |
8XDU
 
 | |
8XDR
 
 | | O-methyltransferase from Lycoris aurea complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDV
 
 | |
