6SFK
 
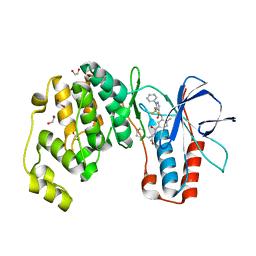 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5YBJ
 
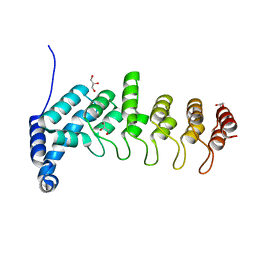 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
6Y7H
 
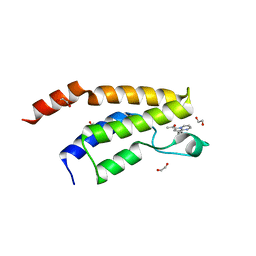 | | Structure of the BRD9 bromodomain and compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-phenylpyrrolo[1,2-a]pyrimidin-6-yl)ethanone, Bromodomain-containing protein 9 | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
6Y7G
 
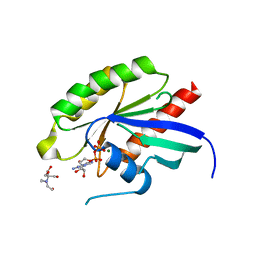 | | Structure of the human RAB3C in complex with GDP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, Raux, B, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human RAB3C in complex with GDP
To Be Published
|
|
6Y7I
 
 | | Structure of the BRD9 bromodomain and compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-pyridin-2-ylpyrrolo[1,2-a]pyrimidin-6-yl)ethanone, Bromodomain-containing protein 9 | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain and compound 2
To Be Published
|
|
6YZ4
 
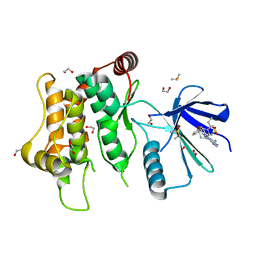 | | Crystal structure of MKK7 (MAP2K7) with ibrutinib bound at allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-06 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6T28
 
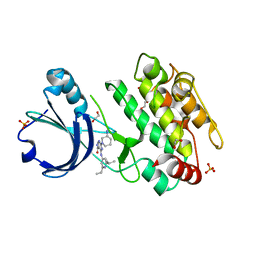 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
5YBV
 
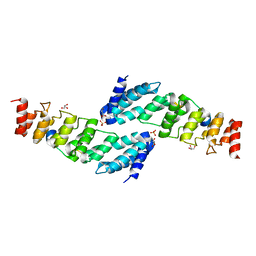 | | The structure of the KANK2 ankyrin domain with the KIF21A peptide | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 2, Kinesin-like protein KIF21A, ... | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
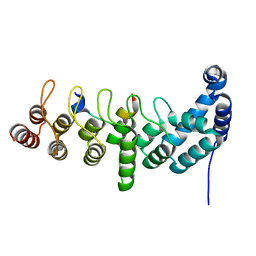 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | Descriptor: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
6YWK
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6T6A
 
 | | Crystal structure of DYRK1A complexed with KuFal319 (compound 11) | | Descriptor: | 4-chloranyl-5~{H}-cyclohepta[b]indol-10-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | [ b ]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors.
Molecules, 24, 2019
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6TSG
 
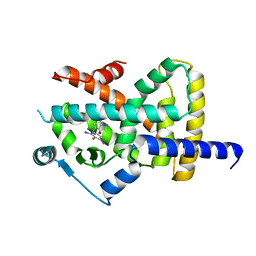 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
6SYP
 
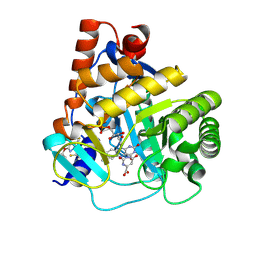 | | Human DHODH bound to inhibitor IPP/CNRS-A017 | | Descriptor: | 2-[4-[2,6-bis(fluoranyl)phenoxy]-5-methyl-3-propan-2-yloxy-pyrazol-1-yl]-5-cyclopropyl-3-fluoranyl-pyridine, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kraemer, A, Janin, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor
Eur.J.Med.Chem., 208, 2020
|
|
6XKB
 
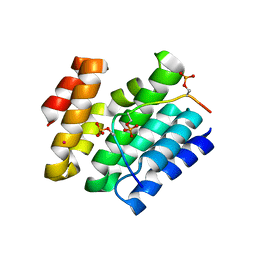 | | Crystal structure of SR-related and CTD-associated factor 4(SCAF4-CID)with peptide S2,S5p-CTD | | Descriptor: | S2,S5p-CTD peptide, SR-related and CTD-associated factor 4, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Febs Lett., 596, 2022
|
|
6RAA
 
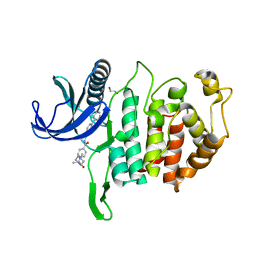 | | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Arrowsmith, C, Knapp, S, Bountra, C, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003
To Be Published
|
|
8BB5
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with Aryl linker | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8BB4
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with C3 linker | | Descriptor: | Elongin-B, Elongin-C, WD repeat-containing protein 5, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8BIN
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Back-Pocket Optimization of 2-Aminopyrimidine-Based Macrocycles Leads to Potent EPHA2/GAK Kinase Inhibitors.
J.Med.Chem., 2024
|
|
8BIO
 
 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MRAL5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-5-(trifluoromethyl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Lucic, A, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
8EM8
 
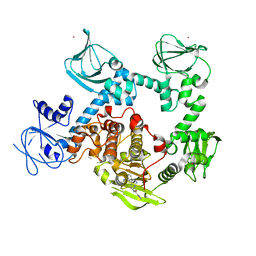 | | Co-crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum in complex with RY-1-165 | | Descriptor: | UNKNOWN ATOM OR ION, [(3R)-3-{[(4M)-4-(4-cyclopropyl-2-phenyl-1H-imidazol-1-yl)pyrimidin-2-yl]amino}pyrrolidin-1-yl](1,3-thiazol-2-yl)methanone, cGMP-dependent protein kinase, ... | | Authors: | Hutchinson, A, Dong, A, Seitova, A, Bhanot, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure-Activity Relationship of a Pyrrole Based Series of PfPKG Inhibitors as Anti-Malarials.
J.Med.Chem., 67, 2024
|
|
3CET
 
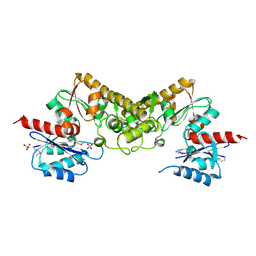 | | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | Conserved archaeal protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, J.F, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
To be Published
|
|
3CEW
 
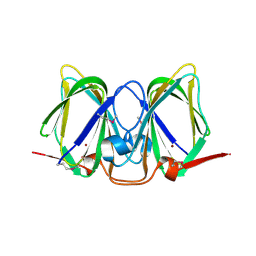 | | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205 | | Descriptor: | Uncharacterized cupin protein, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Wang, H, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of a cupin protein (BF4112) from Bacteroides fragilis. Northeast Structural Genomics Consortium target BfR205.
To be Published
|
|
