7D8U
 
 | | Crystal structure of the C-terminal domain of pNP868R from African swine fever virus | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GTP--RNA guanylyltransferase, ... | | Authors: | Du, X, Geng, Z, Zhang, H. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Biochemical Characteristic of the Methyltransferase (MTase) Domain of RNA Capping Enzyme from African Swine Fever Virus.
J.Virol., 2020
|
|
7DJT
 
 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | Descriptor: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2020-11-21 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
7E5N
 
 | | crystal structure of cis assembled TROP-2 | | Descriptor: | Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7E5M
 
 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
5XW2
 
 | |
8A2C
 
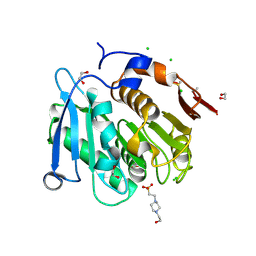 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
8RT0
 
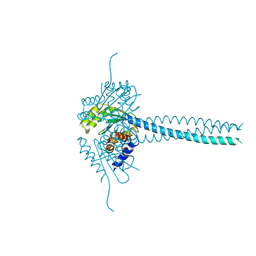 | | BTV-15 VP5 pH 6.0 | | Descriptor: | 1,2-ETHANEDIOL, Outer capsid protein VP5 | | Authors: | Stuart, D.I, Sutton, G.C. | | Deposit date: | 2024-01-25 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The effect of pH on the structure of Bluetongue virus VP5.
J.Gen.Virol., 105, 2024
|
|
8RT1
 
 | | BTV15 VP5 at pH 9.0 | | Descriptor: | Outer capsid protein VP5 | | Authors: | Sutton, G.C, Stuart, D.I. | | Deposit date: | 2024-01-25 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The effect of pH on the structure of Bluetongue virus VP5.
J.Gen.Virol., 105, 2024
|
|
5K24
 
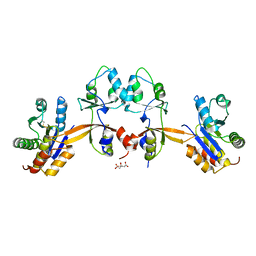 | |
5K25
 
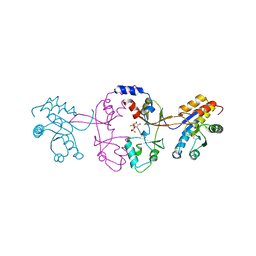 | |
6PU7
 
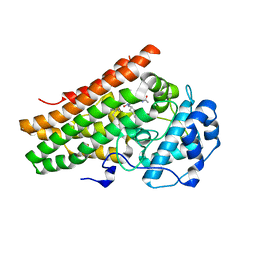 | | Human IDO1 in complex with compound 17 (N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide) | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-{2-[(4-{N-[(7S)-4-fluorobicyclo[4.2.0]octa-1,3,5-trien-7-yl]-N'-hydroxycarbamimidoyl}-1,2,5-oxadiazol-3-yl)sulfanyl]ethyl}acetamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of Amino-cyclobutarene-derived Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitors for Cancer Immunotherapy.
Acs Med.Chem.Lett., 10, 2019
|
|
4RT4
 
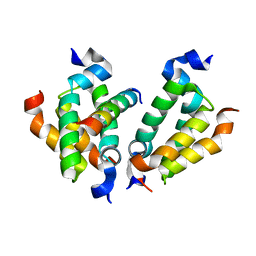 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTA
 
 | |
6UCG
 
 | |
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | Descriptor: | CHLORIDE ION, SULFATE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8X5D
 
 | |
7JMN
 
 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
1RCS
 
 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
6CKN
 
 | | Crystal structure of an AF10 fragment | | Descriptor: | Protein AF-10, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
7FJ2
 
 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
5YAT
 
 | |
3BC5
 
 | | X-ray crystal structure of human ppar gamma with 2-(5-(3-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)benzyl)-2-phenyl-2h-1,2,3-triazol-4-yl)acetic acid | | Descriptor: | (5-{3-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]benzyl}-2-phenyl-2H-1,2,3-triazol-4-yl)acetic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2007-11-12 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, synthesis and structure-activity relationships of azole acids as novel, potent dual PPAR alpha/gamma agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4KBQ
 
 | | Structure of the CHIP-TPR domain in complex with the Hsc70 Lid-Tail domains | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock cognate 71 kDa protein | | Authors: | Page, R.C, Amick, J, Nix, J.C, Misra, S. | | Deposit date: | 2013-04-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Bipartite Interaction between Hsp70 and CHIP Regulates Ubiquitination of Chaperoned Client Proteins.
Structure, 23, 2015
|
|
