4RMW
 
 | |
4Z2B
 
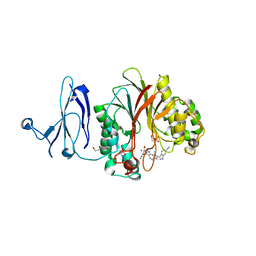 | | The structure of human PDE12 residues 161-609 in complex with GSK3036342A | | Descriptor: | 1,2-ETHANEDIOL, 2',5'-phosphodiesterase 12, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4TS6
 
 | |
4RMU
 
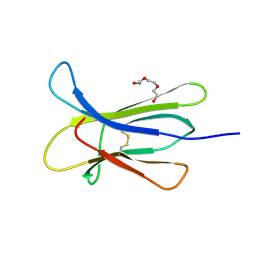 | |
4RMV
 
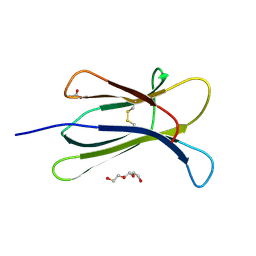 | |
4Z0V
 
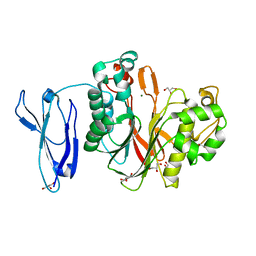 | | The structure of human PDE12 residues 161-609 | | Descriptor: | 2',5'-phosphodiesterase 12, GLYCEROL, MAGNESIUM ION | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-26 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
3G5O
 
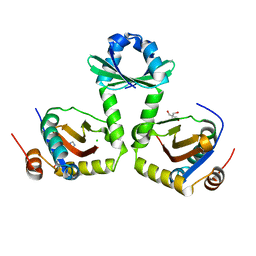 | | The crystal structure of the toxin-antitoxin complex RelBE2 (Rv2865-2866) from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
6MO5
 
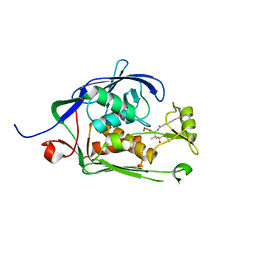 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6MOO
 
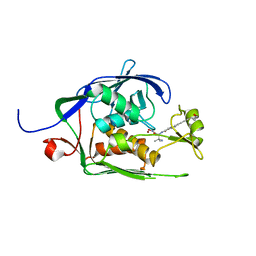 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
8WZ2
 
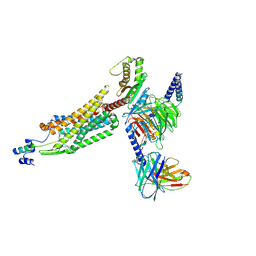 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
6MO4
 
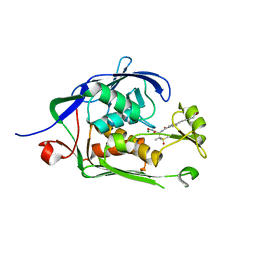 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
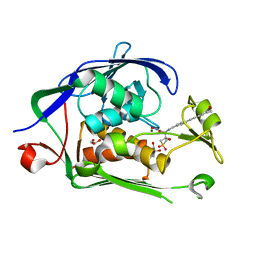 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
2ROG
 
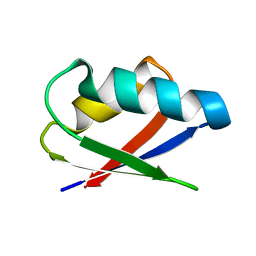 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2RJQ
 
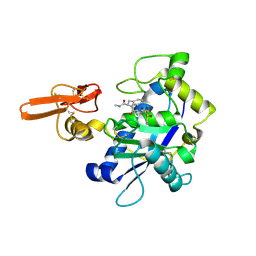 | | Crystal structure of ADAMTS5 with inhibitor bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ADAMTS-5, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2MLB
 
 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MN4
 
 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
7CGW
 
 | | Complex structure of PD-1 and tislelizumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of tislelizumab Fab, Light chain of tislelizumab Fab, ... | | Authors: | Hong, Y, Feng, Y.C, Liu, Y. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tislelizumab uniquely binds to the CC' loop of PD-1 with slow-dissociated rate and complete PD-L1 blockage.
Febs Open Bio, 11, 2021
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | Descriptor: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
8TJL
 
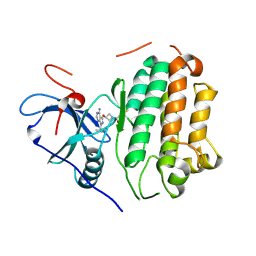 | | EGFR kinase in complex with pyrazolopyrimidine covalent inhibitor | | Descriptor: | 1-{3-[(4-amino-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)oxy]azetidin-1-yl}propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZNL0325, a Pyrazolopyrimidine-Based Covalent Probe, Demonstrates an Alternative Binding Mode for Kinases.
J.Med.Chem., 67, 2024
|
|
3B2Z
 
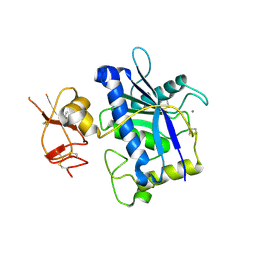 | | Crystal Structure of ADAMTS4 (apo form) | | Descriptor: | ADAMTS-4, CALCIUM ION, ZINC ION | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
6KXA
 
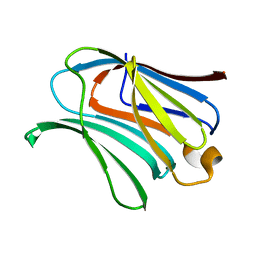 | | Galectin-3 CRD binds to GalA dimer | | Descriptor: | Galectin-3, alpha-D-galactopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid | | Authors: | Su, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Topsy-turvy binding of negatively charged homogalacturonan oligosaccharides to galectin-3.
Glycobiology, 31, 2021
|
|
7DY9
 
 | | Thermotoga maritima ferritin mutant-FLAL | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, X. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
7DY8
 
 | | Thermotoga maritima ferritin mutant-FLAL | | Descriptor: | CALCIUM ION, FE (III) ION, Ferritin | | Authors: | Zhang, X, Zhao, G. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials.
Nat Commun, 12, 2021
|
|
