1RD6
 
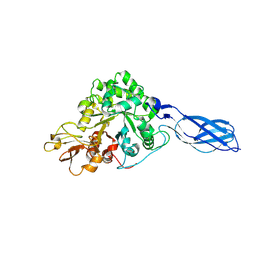 | | Crystal Structure of S. Marcescens Chitinase A Mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation.
Biosci.Biotechnol.Biochem., 70, 2006
|
|
1QWK
 
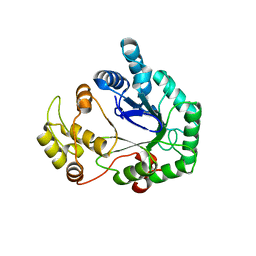 | | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa protein (aldose reductase family member) | | Descriptor: | aldo-keto reductase family 1 member C1 | | Authors: | Chen, L, Zhou, X.E, Meehan, E.J, Symersky, J, Lu, S, Li, S, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-09-02 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa
protein (aldose reductase family member)
To be published
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TDH
 
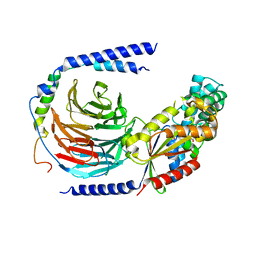 | | The crystal structure of the dominant negative mutant G protein alpha(i)-1-beta-1-gamma-2 G203A/A326S | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, P, Jia, M.-Z, Zhou, X.E, de Waal, P.W, Dickson, B.M, Liu, B, Hou, L, Yin, Y.-T, Kang, Y.-Y, Shi, Y, Melcher, K, Xu, H.E, Jiang, Y. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of the dominant negative phenotype of the G alpha i1 beta 1 gamma 2 G203A/A326S heterotrimer
Acta Pharmacol.Sin., 37, 2016
|
|
5TUB
 
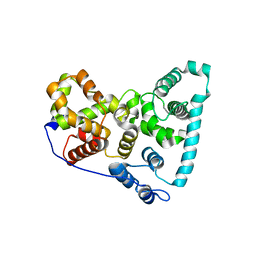 | | Crystal Structure of the Shark TBC1D15 GAP Domain | | Descriptor: | Shark TBC1D15 GTPase-activating Protein | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TUC
 
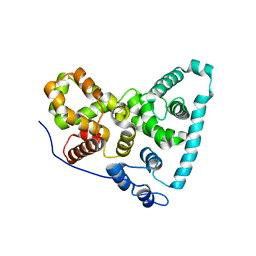 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | Descriptor: | Sus TBC1D15 GAP Domain | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TWO
 
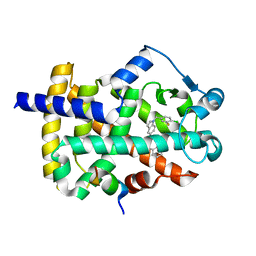 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selectively PPAR gamma-modulating ligand VSP-51 | | Descriptor: | N-benzyl-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-5-carboxamide, PRO-SER-LEU-LEU-LYS-LYS-LEU-LEU-LEU-ALA-PRO, Peroxisome proliferator-activated receptor gamma | | Authors: | Yi, W, Shi, J, Zhao, G, Zhou, X.E, Suino-Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-14 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Identification of a novel selective PPAR gamma ligand with a unique binding mode and improved therapeutic profile in vitro.
Sci Rep, 7, 2017
|
|
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | Authors: | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
6PA7
 
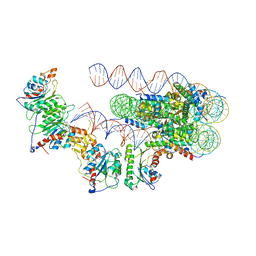 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
6PB1
 
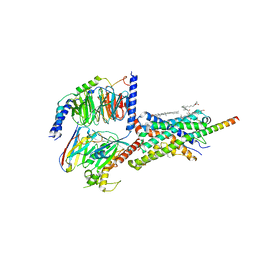 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6PT0
 
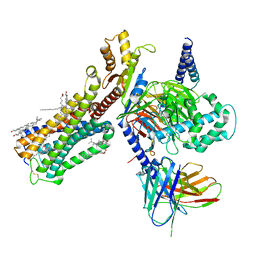 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
6OE8
 
 | | The crystal structure of hyper-thermostable AgUricase mutant K12C/E286C | | Descriptor: | MALONATE ION, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Shi, Y, Wang, T, Zhou, X.E, Liu, Q, Jiang, Y, Xu, H.E. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-based design of a hyperthermostable AgUricase for hyperuricemia and gout therapy.
Acta Pharmacol.Sin., 40, 2019
|
|
6PB0
 
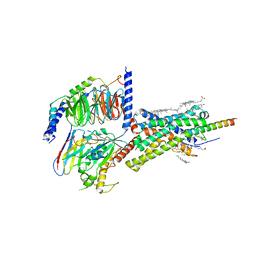 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
7JVQ
 
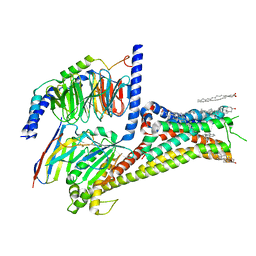 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
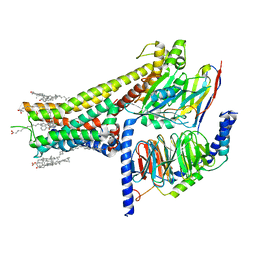 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
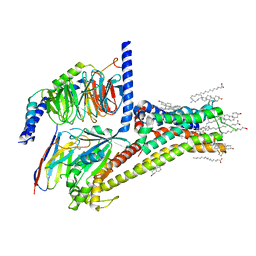 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
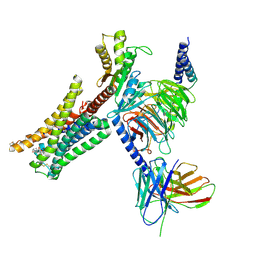 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JHG
 
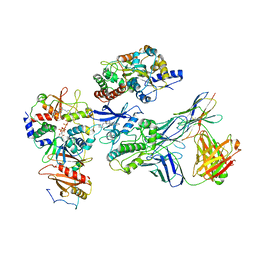 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JHH
 
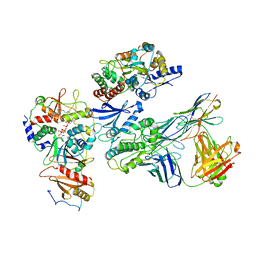 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Fab and nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7JIJ
 
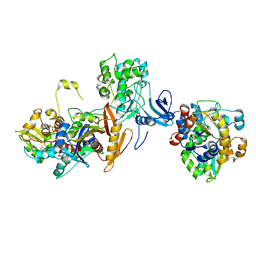 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
3UP0
 
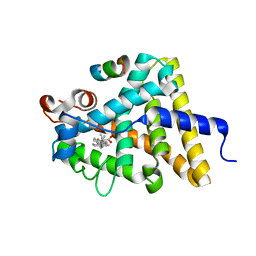 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-delta7-dafachronic acid | | Descriptor: | (5beta,14beta,17alpha,25S)-3-oxocholest-7-en-26-oic acid, Nuclear receptor coactivator 2, aceDAF-12 | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
3UP3
 
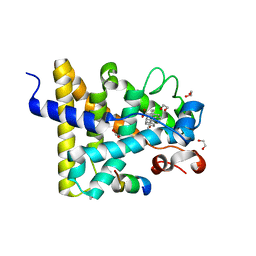 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
