9D65
 
 | | Nitrile hydratase BR52A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Miller, C.G, Holz, R.C, Liu, D, Kaley, N. | | Deposit date: | 2024-08-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Role of second-sphere arginine residues in metal binding and metallocentre assembly in nitrile hydratases.
J Inorg Biochem, 256, 2024
|
|
9D5U
 
 | | Nitrile hydratase S112A mutant | | Descriptor: | Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, GLYCEROL | | Authors: | Miller, C.G, Holz, R.C, Liu, D. | | Deposit date: | 2024-08-14 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Catalytic and post-translational maturation roles of a conserved active site serine residue in nitrile hydratases.
J.Inorg.Biochem., 262, 2024
|
|
4UY9
 
 | | Structure of MLK1 kinase domain with leucine zipper 1 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
4UYA
 
 | | Structure of MLK4 kinase domain with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE MLK4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
2KRR
 
 | | Solution structure of the RBD1,2 domains from human nucleolin | | Descriptor: | Nucleolin | | Authors: | Arumugam, N, Miller, C, Maliekal, J, Bates, P.J, Trent, J.O, Lane, A.N. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RBD1,2 domains from human nucleolin.
J.Biomol.Nmr, 47, 2010
|
|
3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
4R67
 
 | | Human constitutive 20S proteasome in complex with carfilzomib | | Descriptor: | N-{(2S)-2-[(morpholin-4-ylacetyl)amino]-4-phenylbutanoyl}-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-phenylalaninamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
4R3O
 
 | | Human Constitutive 20S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
5JYQ
 
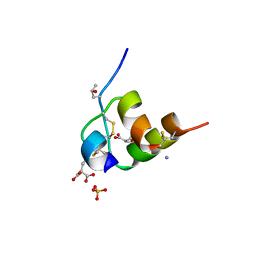 | | Structure of Conus Geographus insulin G1 | | Descriptor: | Insulin 1, Insulin 1b, SULFATE ION | | Authors: | Lawrence, M.C, Menting, J, Norton, R.S, Safavi-Hemami, H. | | Deposit date: | 2016-05-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6CQS
 
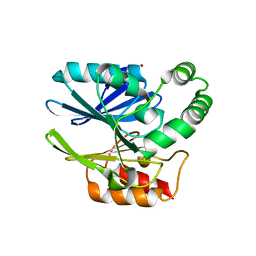 | | Sediminispirochaeta smaragdinae SPS-1 metallo-beta-lactamase | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Page, R.C, VanPelt, J, Cheng, Z, Crowder, M.W. | | Deposit date: | 2018-03-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Metal Center Drives the Activity of the Sediminispirochaeta smaragdinae Metallo-beta-lactamase SPS-1.
Biochemistry, 57, 2018
|
|
8TFT
 
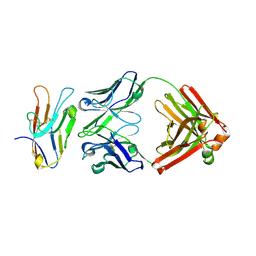 | | Fab of O13-1 human IgG1 antibody bound to IgV domain of human TIM-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O-13-1 IgG1 Fab heavy chain, O-13-1 IgG1 Fab light chain, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Yang, C. | | Deposit date: | 2023-07-11 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation of AZD7789, a novel PD-1 and TIM-3 targeting bispecific antibody, which binds to a differentiated epitope of TIM-3
To be published
|
|
8TBB
 
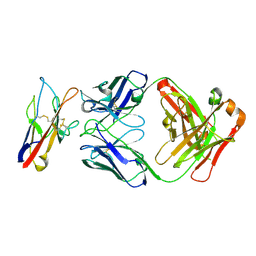 | | F9S, novel TIM-3 targeting antibody, bound to IgV domain of TIM-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F9S Fab heavy chain, F9S Fab light chain, ... | | Authors: | Oganesyan, V, van Dyk, N, Mazor, Y, Yang, C. | | Deposit date: | 2023-06-28 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Generation of AZD7789, a novel PD-1 and TIM-3 targeting bispecific antibody, which binds to a differentiated epitope of TIM-3
To be published
|
|
5UD5
 
 | |
5V6X
 
 | |
8BGS
 
 | |
8BGV
 
 | |
6NPE
 
 | | C-abl Kinase domain with the activator(cmpd6), 2-cyano-N-(4-(3,4-dichlorophenyl)thiazol-2-yl)acetamide | | Descriptor: | 2-cyano-~{N}-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]ethanamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, NONAETHYLENE GLYCOL, ... | | Authors: | campobasso, N. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and Optimization of Novel Small c-Abl Kinase Activators Using Fragment and HTS Methodologies.
J. Med. Chem., 62, 2019
|
|
6NPV
 
 | |
6NPU
 
 | |
5EVG
 
 | | Crystal structure of a Francisella virulence factor FvfA in the orthorhombic form | | Descriptor: | Francisella virulence factor | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5EVF
 
 | | Crystal structure of a Francisella virulence factor FvfA in the hexagonal form | | Descriptor: | CHLORIDE ION, Francisella virulence factor, GLYCEROL | | Authors: | Kolappan, S, Lo, K.Y, Shen, C.L.J, Guttman, J.A, Craig, L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-10-26 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structure of the conserved Francisella virulence protein FvfA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6GYZ
 
 | |
6GYW
 
 | |
6GYY
 
 | |
6GYX
 
 | |
