3KCY
 
 | | Factor inhibiting HIF-1 alpha in complex with 8-hydroxyquinoline | | Descriptor: | FE (II) ION, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Moon, H, Han, S, Choe, J. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of human FIH-1 in complex with quinol family inhibitors
Mol.Cells, 29, 2010
|
|
4HCT
 
 | | Crystal structure of ITK in complex with compound 52 | | Descriptor: | 3-{1-[(3R)-1-acryloylpiperidin-3-yl]-4-amino-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-(3-tert-butylphenyl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Zapf, C.W, Gerstenberger, B.S, Xing, L, Limburg, D.C, Anderson, D.R, Caspers, N, Han, S, Aulabaugh, A, Kurumbail, R, Shakya, S, Li, X, Spaulding, V, Czerwinski, R.M, Seth, N, Medley, Q.G. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
2GM7
 
 | | TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Ten A Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
8JYU
 
 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Acyl-ACP Synthetase structure bound to Decanoyl-AMP
To Be Published
|
|
8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Acyl-ACP Synthetase structure bound to C10-AMS
To Be Published
|
|
8P1U
 
 | | Structure of divisome complex FtsWIQLB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ, ... | | Authors: | Yang, L, Chang, S, Tang, D, Dong, H. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the activation of the divisome complex FtsWIQLB.
Cell Discov, 10, 2024
|
|
2GM8
 
 | | TenA Homolog/Thi-4 Thiaminase complexed with product 4-amino-5-hydroxymethyl-2-methylpyrimidine | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-5-HYDROXYMETHYL-2-METHYLPYRIMIDINE, tenA homolog/Thi-4 Thiaminase | | Authors: | Sawaya, M.R, Chan, S, Han, G.W, Perry, L.J, Pashkov, I. | | Deposit date: | 2006-04-06 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a TenA Homolog/Thi-4 Thiaminase from Pyrobaculum Aerophilum
To be Published
|
|
6M0J
 
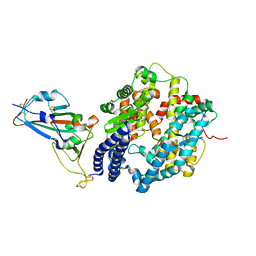 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
8BNZ
 
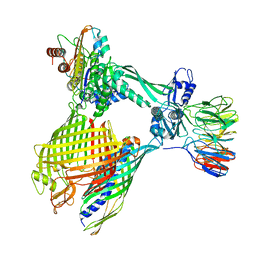 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
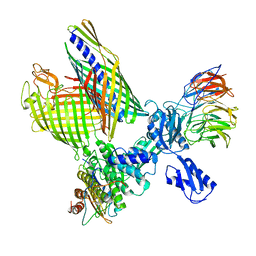 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
7KDP
 
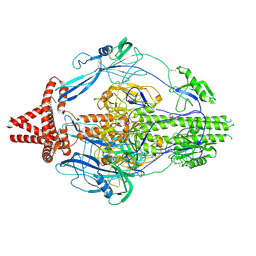 | | HCMV prefusion gB in complex with fusion inhibitor WAY-174865 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
7KDD
 
 | | HCMV postfusion gB in complex with SM5-1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-08 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
2KER
 
 | | alpha-amylase inhibitor Parvulustat (Z-2685) from Streptomyces parvulus | | Descriptor: | Alpha-amylase inhibitor Z-2685 | | Authors: | Rehm, S, Han, S, Hassani, I, Sokocevic, A, Jonker, H.R.A, Engels, J.W, Schwalbe, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The high resolution NMR structure of parvulustat (Z-2685) from Streptomyces parvulus FH-1641: comparison with tendamistat from Streptomyces tendae 4158
Chembiochem, 10, 2009
|
|
2K4U
 
 | | Solution structure of the SCORPION TOXIN ADWX-1 | | Descriptor: | Potassium channel toxin alpha-KTx 3.6 | | Authors: | Yin, S.J, Jiang, L, Yi, H, Han, S, Yang, D.W, Liu, M.L, Liu, H, Cao, Z.J, Wu, Y.L, Li, W.X. | | Deposit date: | 2008-06-18 | | Release date: | 2008-12-09 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Different Residues in Channel Turret Determining the Selectivity of ADWX-1 Inhibitor Peptide between Kv1.1 and Kv1.3 Channels
J.Proteome Res., 7, 2008
|
|
1WWJ
 
 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
1YC4
 
 | | Crystal structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1H-IMIDAZOL-4-YL)-3-(5-ETHYL-2,4-DIHYDROXY-PHENYL)-1H-PYRAZOLE, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC3
 
 | | Crystal Structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC1
 
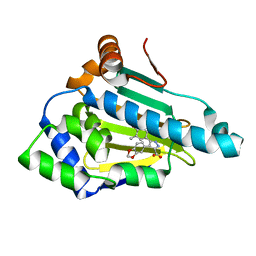 | | Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3G12
 
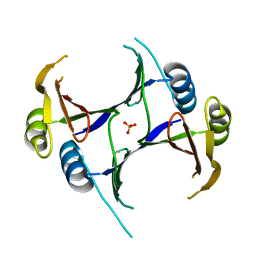 | | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus | | Descriptor: | Putative lactoylglutathione lyase, SULFATE ION | | Authors: | Patskovsky, Y, Madegowda, M, Gilmore, M, Chang, S, Maletic, M, Smith, D, Sauder, J.M, Burley, S.K, Swaminathan, S, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of a putative lactoylglutathione lyase from Bdellovibrio bacteriovorus
To be Published
|
|
6VGI
 
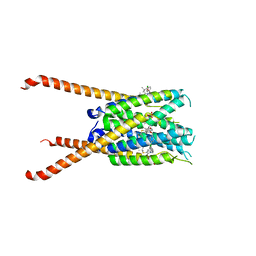 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
7X9B
 
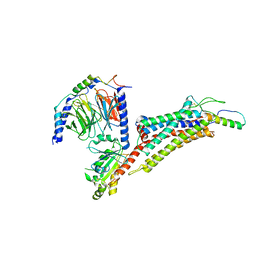 | | Cryo-EM structure of neuropeptide Y Y2 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7X9A
 
 | | Cryo-EM structure of neuropeptide Y Y1 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7X9C
 
 | | Cryo-EM structure of neuropeptide Y Y4 receptor in complex with PP and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
