8WF4
 
 | | The Crystal Structure of RSK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of RSK1 from Biortus.
To Be Published
|
|
8WFY
 
 | | The Crystal Structure of SHP2 from Biortus. | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of SHP2 from Biortus.
To Be Published
|
|
4JCL
 
 | | Crystal structure of Alpha-CGT from Paenibacillus macerans at 1.7 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J, Wu, J, Li, J, Chen, J. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Alpha-Cgt from Paenibacillus Macerans at 1.7 Angstrom Resolution
To be Published
|
|
4WQN
 
 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
2ZKJ
 
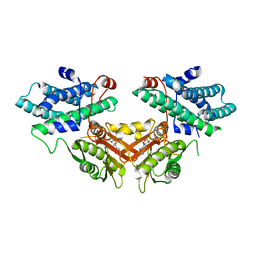 | | Crystal structure of human PDK4-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Li, J, Chuang, D.T. | | Deposit date: | 2008-03-25 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity
J.Biol.Chem., 283, 2008
|
|
1TBU
 
 | |
4LAD
 
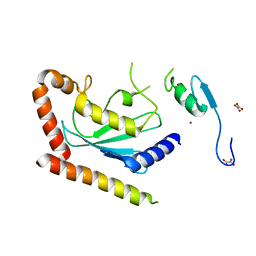 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
7C84
 
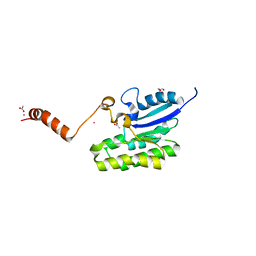 | | Esterase AlinE4 mutant, D162A | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
5EJO
 
 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
4BGD
 
 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, T.H.D, Li, J, Nagai, K. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
7C85
 
 | | Esterase AlinE4 mutant-S13A | | Descriptor: | ACETATE ION, CADMIUM ION, SGNH-hydrolase family esterase | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
1TR4
 
 | | Solution structure of human oncogenic protein gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Yuan, C, Li, J, Mahajan, A, Poi, M.J, Byeon, I.J, Tsai, M.D. | | Deposit date: | 2004-06-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship.
Biochemistry, 43, 2004
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | Descriptor: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | Authors: | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8J1I
 
 | | Crystal Structure of EphA8/SASH1 Complex | | Descriptor: | Ephrin type-A receptor 8, SAM and SH3 domain-containing protein 1 | | Authors: | Liu, W, Li, J, Ding, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SASH1: A Novel Eph Receptor Partner and Insights into SAM-SAM Interactions.
J.Mol.Biol., 435, 2023
|
|
8Y4G
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zeng, P, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-01-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8Y4H
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Guo, L, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-01-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
3L3Z
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
3L3X
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
8DSV
 
 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ8
 
 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
 | |
6V4P
 
 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
8YBQ
 
 | | Choline transporter BetT - CHT bound | | Descriptor: | BCCT family transporter, CHOLINE ION | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
