3JVN
 
 | | Crystal Structure of the acetyltransferase VF_1542 from Vibrio fischeri, Northeast Structural Genomics Consortium Target VfR136 | | Descriptor: | Acetyltransferase, SULFATE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Northeast Structural Genomics Consortium Target VfR136
To be Published
|
|
3K6A
 
 | |
3JZY
 
 | | Crystal structure of human Intersectin 2 C2 domain | | Descriptor: | Intersectin 2, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tempel, W, Tong, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of human Intersectin 2 C2 domain
To be Published
|
|
3K9L
 
 | | Allosteric modulation of H-Ras GTPase | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Fetics, S, Young, M, Buhrman, G, Mattos, C. | | Deposit date: | 2009-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric modulation of H-Ras GTPase
To be Published
|
|
3KAL
 
 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3J3V
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3KAK
 
 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3J9L
 
 | | Structure of Dark apoptosome from Drosophila melanogaster | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
3JXS
 
 | | Crystal structure of XG34, an evolved xyloglucan binding CBM | | Descriptor: | ACETATE ION, CALCIUM ION, Xylanase | | Authors: | Divne, C, Tan, T.-C, Brumer, H, Gullfot, F. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module.
Proteins, 78, 2009
|
|
3JCM
 
 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
3JR7
 
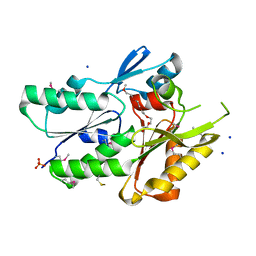 | | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149
To be Published
|
|
3K1K
 
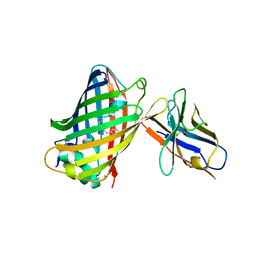 | | Green fluorescent protein bound to enhancer nanobody | | Descriptor: | Enhancer, Green Fluorescent Protein | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-09-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K1N
 
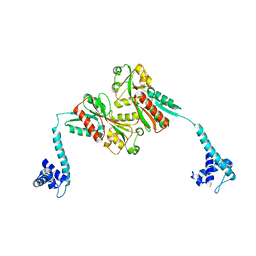 | |
3K4N
 
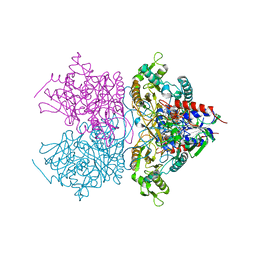 | | Pyranose 2-oxidase F454A/S455A/Y456A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Divne, C, Tan, T.C. | | Deposit date: | 2009-10-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
Febs J., 277, 2010
|
|
3K51
 
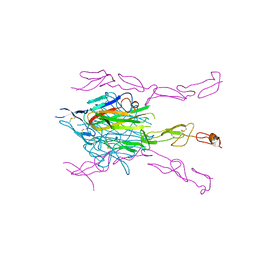 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3K7Y
 
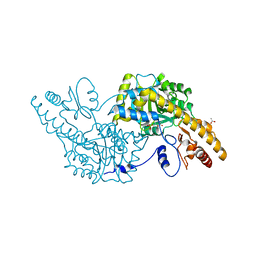 | | Aspartate Aminotransferase of Plasmodium falciparum | | Descriptor: | ACETATE ION, Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Groves, M.R, Jordanova, R, Jain, R, Wrenger, C, Muller, I.B. | | Deposit date: | 2009-10-13 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Inhibition of the Aspartate Aminotransferase of Plasmodium falciparum.
J.Mol.Biol., 405, 2011
|
|
9F59
 
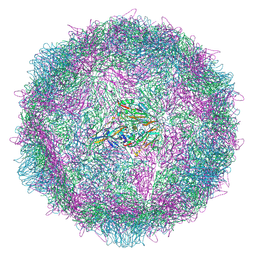 | | Poliovirus type 2 (strain MEF-1) stabilised virus-like particle (PV2 SC6b) from a mammalian expression system. | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-28 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
3K7K
 
 | | Crystal structure of the complex between Carbonic Anhydrase II and anions | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, ZINC ION, ... | | Authors: | Temperini, C. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. X-ray crystal studies of the carbonic anhydrase II-trithiocarbonate adduct-An inhibitor mimicking the sulfonamide and urea binding to the enzyme
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K9R
 
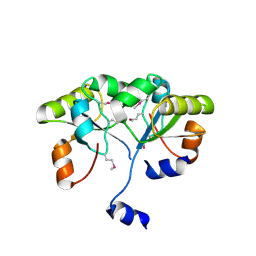 | | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp. Northeast Structural Genomics Consortium Target NsR437c. | | Descriptor: | Alr3790 protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of the Rhodanese-like domain of the Alr3790 protein from Anabaena sp.
To be Published
|
|
3KAO
 
 | | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SULFATE ION, Tagatose 1,6-diphosphate aldolase, ... | | Authors: | Chang, C, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus
To be Published
|
|
9F0K
 
 | | Poliovirus type 1 (strain Mahoney) expanded conformation stabilised virus-like particle (PV1 SC6b) from a mammalian expression system | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-16 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
3KAV
 
 | | Crystal Structure of THE MUTANT (L80M) PA2107 PROTEIN from Pseudomonas aeruginosa, Northeast Structural Genomics Consortium Target PaR198 | | Descriptor: | uncharacterized protein PA2107 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target PaR198
To be Published
|
|
3J8Y
 
 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
9F5P
 
 | | Poliovirus type 2 (strain MEF-1) stabilised virus-like particle (PV2 SC6b) from an insect cell expression system. | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-29 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9F7O
 
 | |
