6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
7WNM
 
 | | Structure of SARS-CoV-2 Gamma variant receptor-binding domain complexed with high affinity human ACE2 mutant (T27F,R273Q) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Ma, R.Y, Han, P.C, Wang, Q.H, Han, P. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A binding-enhanced but enzymatic activity-eliminated human ACE2 efficiently neutralizes SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
2MMS
 
 | | AG(7-deaza)G FAPY modified duplex | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*(7GU)P*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*CP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
5X26
 
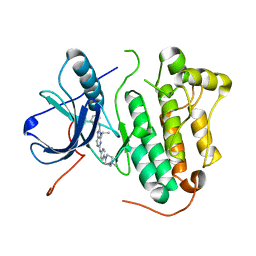 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2K
 
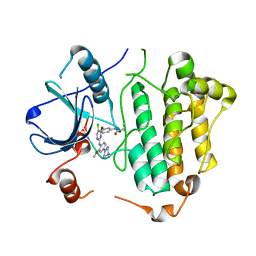 | | Crystal structure of EGFR 696-1022 T790M in complex with WZ4003 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Zhu, S.J, Zhao, P, Yun, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5XGM
 
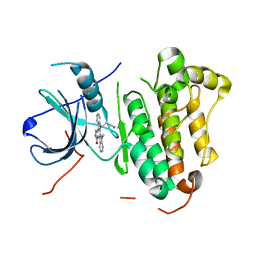 | | Crystal structure of EGFR 696-1022 T790M in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5X28
 
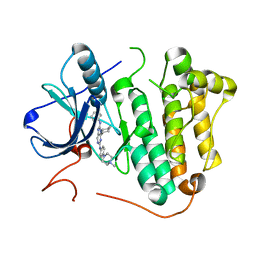 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2C
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(5) | | Descriptor: | 1,2-ETHANEDIOL, 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, ... | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2F
 
 | | Crystal structure of EGFR 696-1022 T790M/V948R in complex with SKLB(6) | | Descriptor: | 9-cyclohexyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X27
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(5) | | Descriptor: | 9-cyclopentyl-N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-purine-2,8-diamine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yun, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5YJ8
 
 | |
5XZU
 
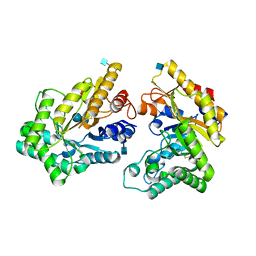 | | Crystal structure of GH10 xylanase from Bispora. sp MEY-1 with xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | You, S, Chen, C.C, Tu, T, Guo, R.T, Luo, H.Y, Yao, B. | | Deposit date: | 2017-07-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into the functional roles of Glu175 in the hyperthermostable xylanase XYL10C-Delta N through structural analysis and site-saturation mutagenesis.
Biotechnol Biofuels, 11, 2018
|
|
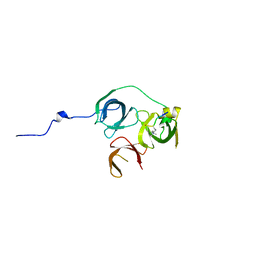
5Y5W
 
 | |
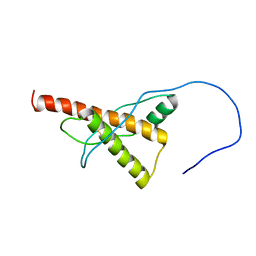
5YJ5
 
 | |
8WZB
 
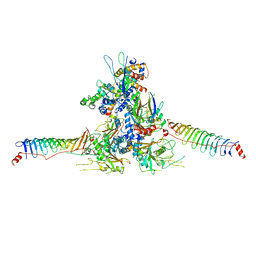 | | RS head-neck monomer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
8X2U
 
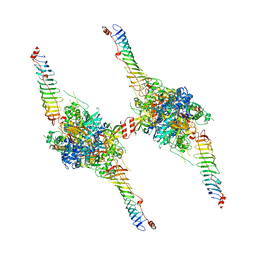 | | Radial spoke head-neck dimer | | Descriptor: | DPY30 domain containing 2, DnaJ homolog subfamily B member 13, Nucleoside diphosphate kinase homolog 5, ... | | Authors: | Meng, X, Cong, Y. | | Deposit date: | 2023-11-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Multi-scale structures of the mammalian radial spoke and divergence of axonemal complexes in ependymal cilia.
Nat Commun, 15, 2024
|
|
5X2A
 
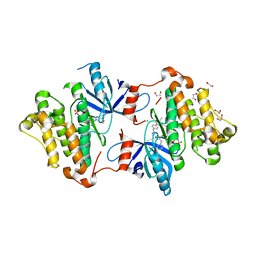 | |
5YJ4
 
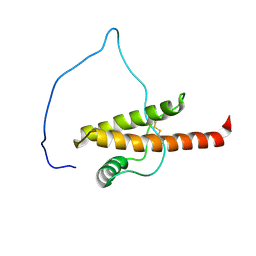 | |
5XGN
 
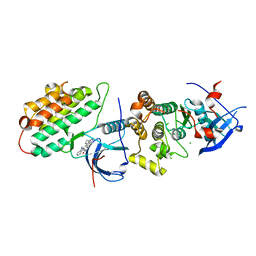 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
4JWH
 
 | | Crystal structure of spTrm10(Full length)-SAH complex | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWJ
 
 | | Crystal structure of scTrm10(84)-SAH complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWG
 
 | | Crystal structure of spTrm10(74) | | Descriptor: | ACETIC ACID, tRNA (guanine(9)-N1)-methyltransferase | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
2MMF
 
 | | Solution structure of AGA modified | | Descriptor: | DNA_(5'-D(*CP*TP*AP*AP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA_(5'-D(*TP*GP*AP*AP*TP*CP*TP*TP*AP*G)-3') | | Authors: | Li, L, Stone, M. | | Deposit date: | 2014-03-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct.
Chem.Res.Toxicol., 28, 2015
|
|
