6KX2
 
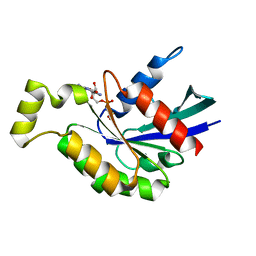 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
8FAD
 
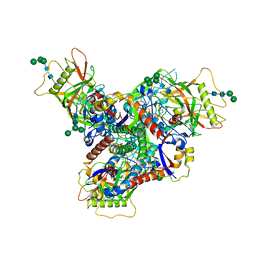 | | Asymmetric structure of cleaved HIV-1 AD8 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
8FAE
 
 | | Asymmetric structure of cleaved HIV-1 AE2 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles (AE2.1) | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
5WUU
 
 | |
7N6W
 
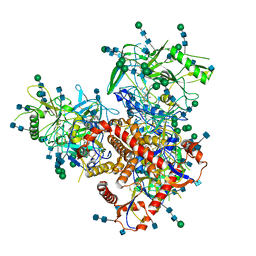 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
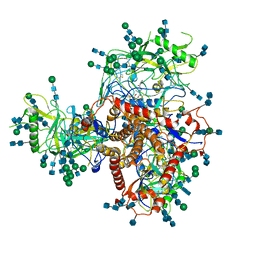 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
5LOV
 
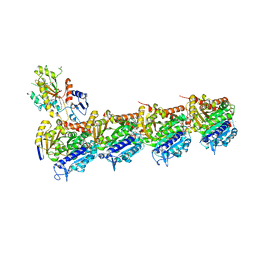 | | DZ-2384 tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DZ 2384, ... | | Authors: | Prota, A.E, Steinmetz, M.O, Shore, G.C, Brouhard, G, Roulston, A. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The synthetic diazonamide DZ-2384 has distinct effects on microtubule curvature and dynamics without neurotoxicity.
Sci Transl Med, 8, 2016
|
|
3NSQ
 
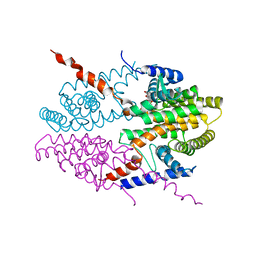 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3NSP
 
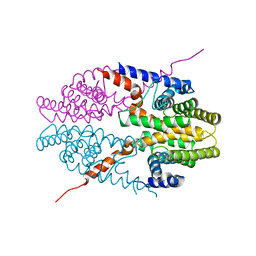 | | Crystal structure of tetrameric RXRalpha-LBD | | Descriptor: | Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5H21
 
 | | Trimethoxy-ring inhibitor in complex with the first bromodomain of BRD4 | | Descriptor: | 3,4,5-trimethoxy-~{N}-(2-thiophen-2-ylethyl)benzamide, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of novel trimethoxy-ring BRD4 bromodomain inhibitors: AlphaScreen assay, crystallography and cell-based assay.
Medchemcomm, 8, 2017
|
|
4FD5
 
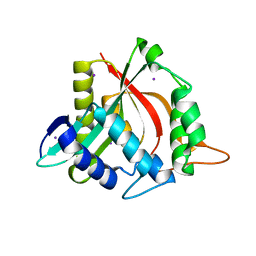 | |
4FD4
 
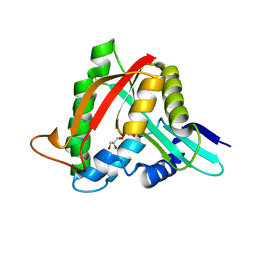 | |
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
1TFT
 
 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | 1-[3,3-DIMETHYL-2-(2-METHYLAMINO-PROPIONYLAMINO)-BUTYRYL]-4-PHENOXY-PYRROLIDINE-2-CARBOXYLIC ACID(1,2,3,4-TETRAHYDRO-NAPHTHALEN-1-YL)-AMIDE, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A.K, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer.
J.Med.Chem., 47, 2004
|
|
1TFQ
 
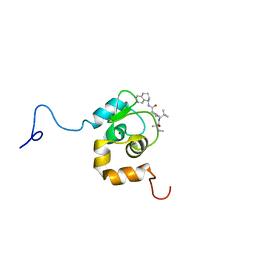 | | NMR Structure of an Antagonists of the XIAP-Caspase-9 Interaction Complexed to the BIR3 domain of XIAP | | Descriptor: | Baculoviral IAP repeat-containing protein 4, N-METHYLALANYL-3-METHYLVALYL-N-(1,2,3,4-TETRAHYDRONAPHTHALEN-1-YL)PROLINAMIDE, ZINC ION | | Authors: | Oost, T.K, Sun, C, Armstrong, R.C, Al-Assaad, A.S, Betz, S.F, Deckwerth, T.L, Elmore, S.W, Meadows, R.P, Olejniczak, E.T, Oleksijew, A, Oltersdorf, T, Rosenberg, S.H, Shoemaker, A.R, Zou, H, Fesik, S.W. | | Deposit date: | 2004-05-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Discovery of Potent Antagonists of the Antiapoptotic Protein XIAP for the Treatment of Cancer.
J.Med.Chem., 47, 2004
|
|
1PGV
 
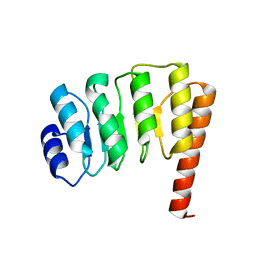 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
4LXD
 
 | | Bcl_2-Navitoclax Analog (without Thiophenyl) Complex | | Descriptor: | 4-(4-{[4-(4-chlorophenyl)-5,6-dihydro-2H-pyran-3-yl]methyl}piperazin-1-yl)-N-{[3-nitro-4-(tetrahydro-2H-pyran-4-ylamino)phenyl]sulfonyl}benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
4LVT
 
 | | Bcl_2-Navitoclax (ABT-263) Complex | | Descriptor: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
4MAN
 
 | | Bcl_2-Navitoclax Analog (with Indole) Complex | | Descriptor: | 4-[4-({4'-chloro-3-[2-(dimethylamino)ethoxy]biphenyl-2-yl}methyl)piperazin-1-yl]-2-(1H-indol-5-yloxy)-N-({3-nitro-4-[(tetrahydro-2H-pyran-4-ylmethyl)amino]phenyl}sulfonyl)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Park, C.H. | | Deposit date: | 2013-08-16 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets.
NAT.MED. (N.Y.), 19, 2013
|
|
5Y1Y
 
 | |
5Z5T
 
 | | The first bromodomain of BRD4 with compound BDF-2141 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5V
 
 | | The first bromodomain of BRD4 with compound BDF-1253 | | Descriptor: | Bromodomain-containing protein 4, N-{8-hydroxy-4-[(1H-imidazol-1-yl)methyl]quinolin-2-yl}acetamide | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
5Z5U
 
 | | The first bromodomain of BRD4 with compound BDF-2254 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinoline-6,8-diol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
