8ZRU
 
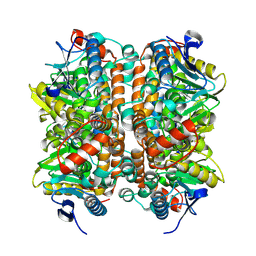 | | Structure of human ECHS1 in apo state | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRV
 
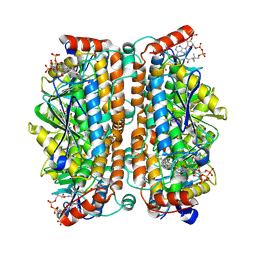 | | Structure of human ECHS1 in complex with Hexanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, HEXANOYL-COENZYME A, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRX
 
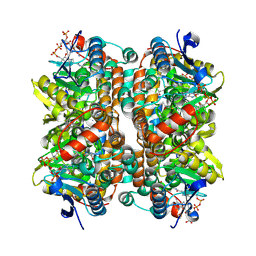 | | Structure of human ECHS1 in complex with Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase, mitochondrial, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRW
 
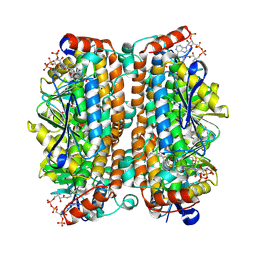 | | Structure of human ECHS1 in complex with Octanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
4V94
 
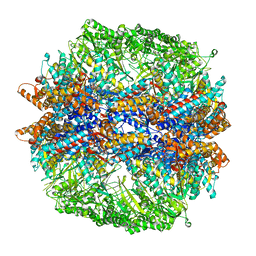 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
5JYN
 
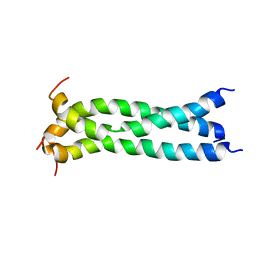 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3S5A
 
 | | ABH2 cross-linked to undamaged dsDNA-2 with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*CP*AP*CP*TP*GP*TP*CP*G)-3', 5'-D(*TP*CP*GP*AP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Ramirez, B, Zhang, W, Jia, G, Zhang, L, Li, C.Q, Dinner, A.R, Yang, C.-G, He, C. | | Deposit date: | 2011-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3S57
 
 | | ABH2 cross-linked with undamaged dsDNA-1 containing cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*TP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Ramirez, B, Zhang, W, Jia, G, Zhang, L, Li, C.Q, Dinner, A.R, Yang, C.-G, He, C. | | Deposit date: | 2011-05-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
6L2P
 
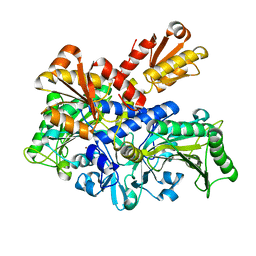 | |
6L2Q
 
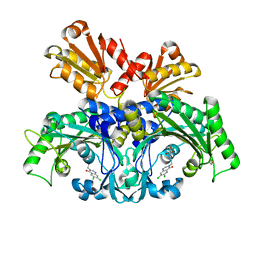 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2019-10-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel tRNA-amino acid dual-site inhibitors against threonyl-tRNA synthetase by fragment-based target hopping.
Eur.J.Med.Chem., 187, 2019
|
|
3RZL
 
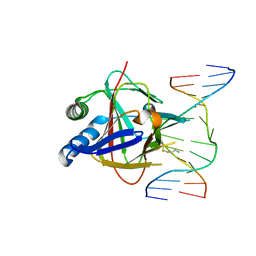 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*CP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*IP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
9LOT
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with N-piperidine ibrutinib | | Descriptor: | 3-(4-phenoxyphenyl)-1-piperidin-4-yl-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORZOXAZONE, SULFATE ION, ... | | Authors: | Peng, X, Chen, B, Xia, K, Zhou, H. | | Deposit date: | 2025-01-23 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9LPD
 
 | | Crystal structure of Escherichia coli trptophanyl-tRNA synthetase in complex with tirabrutinib | | Descriptor: | CHLORZOXAZONE, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Peng, X, Chen, B, Xia, K, Zhou, H. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The lineage-specific tRNA recognition mechanism of bacterial trptophanyl-tRNA synthetase and its implications for inhibitor discover
To Be Published
|
|
9LPC
 
 | |
6UJV
 
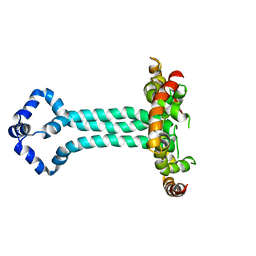 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
8HNI
 
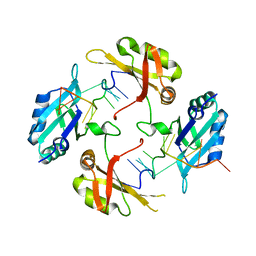 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
6UJU
 
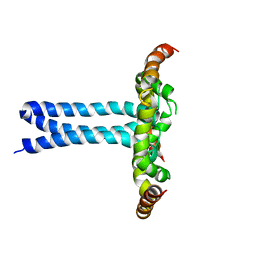 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
7USD
 
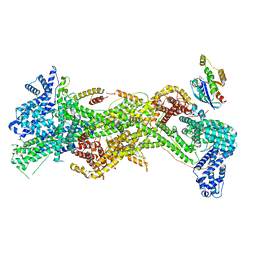 | | Cryo-EM structure of D-site Rac1-bound WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USE
 
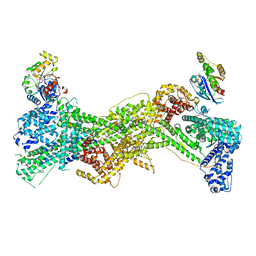 | | Cryo-EM structure of WAVE regulatory complex with Rac1 bound on both A and D site | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USC
 
 | | Cryo-EM structure of WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, Nck-associated protein 1, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
8D56
 
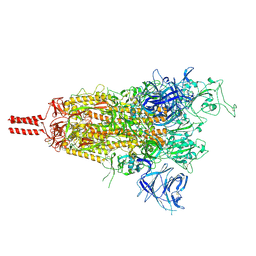 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
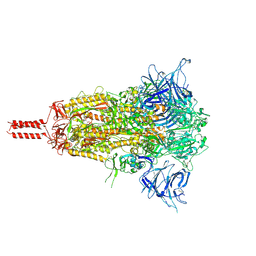 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5C5B
 
 | | Crystal Structure of Human APPL BAR-PH Heterodimer | | Descriptor: | DCC-interacting protein 13-alpha, DCC-interacting protein 13-beta, GLYCEROL | | Authors: | Chen, Y.J, Chen, B. | | Deposit date: | 2015-06-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human APPL BAR-PH Heterodimer
To Be Published
|
|
5L1N
 
 | | Pyrococcus horikoshii CoA Disulfide Reductase Quadruple Mutant | | Descriptor: | COENZYME A, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Sea, K, Chen, B, Crane III, E.J, Sazinsky, M.H. | | Deposit date: | 2016-07-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A broader active site inPyrococcus horikoshiiCoA disulfide reductase accommodates larger substrates and reveals evidence of subunit asymmetry.
FEBS Open Bio, 8, 2018
|
|
