7QKN
 
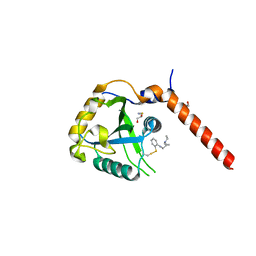 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 7 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7QL7
 
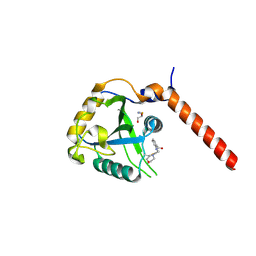 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 9 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
8P29
 
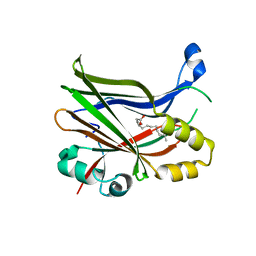 | | TEAD2 in complex with an inhibitor | | Descriptor: | 5-methyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Guichou, J.F, Gelin, M, Allemand, F. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8G4A
 
 | |
8K8T
 
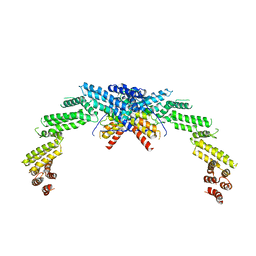 | | Structure of CUL3-RBX1-KLHL22 complex | | Descriptor: | Cullin-3, Kelch-like protein 22 | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8P9Y
 
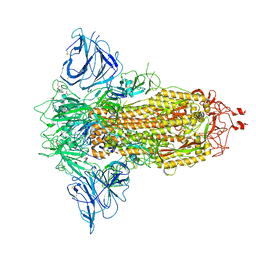 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
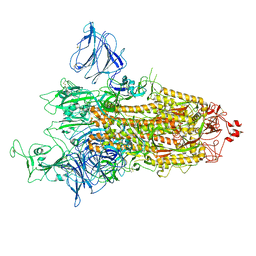 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8POJ
 
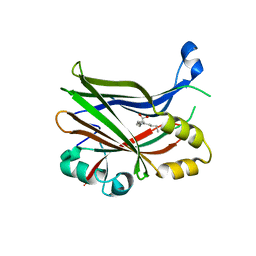 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8PON
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POM
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8BXA
 
 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
1UNL
 
 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | Descriptor: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNG
 
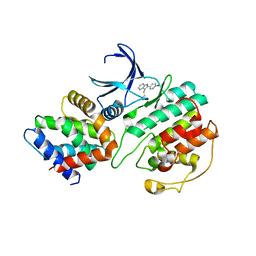 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
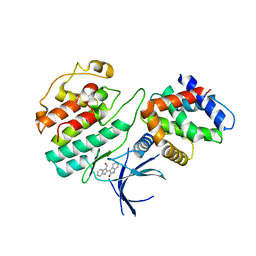 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
371D
 
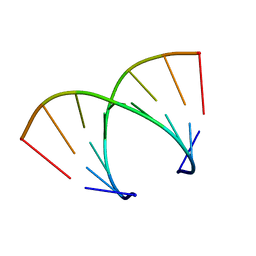 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
370D
 
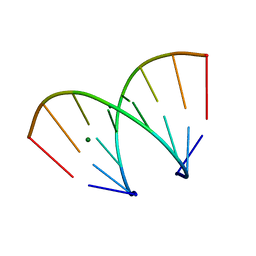 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
369D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
6SD9
 
 | | Crystal structure of wild-type cMET bound by foretinib | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDC
 
 | | Crystal structure of D1228V cMET bound by foretinib | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDD
 
 | | Crystal structure of D1228V cMET bound by BMS-777607 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-{4-[(2-amino-3-chloropyridin-4-yl)oxy]-3-fluorophenyl}-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDE
 
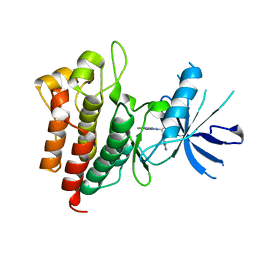 | |
6USY
 
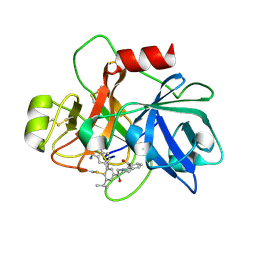 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
8B5G
 
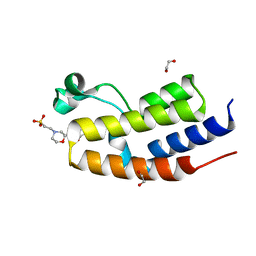 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 7,8-dimethoxy-3-methyl-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,8-dimethoxy-3-methyl-1~{H}-3-benzazepin-2-one, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
8B5H
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH (R)-7-((R)-1,2-dihydroxyethyl)-1,3-dimethyl-5-(1-methyl-1H-pyrazol-4-yl)-1,3-dihydro-2H-benzo[d]azepin-2-one | | Descriptor: | (1~{R})-7-[(1~{R})-1,2-bis(oxidanyl)ethyl]-1,3-dimethyl-5-(1-methylpyrazol-4-yl)-1~{H}-3-benzazepin-2-one, 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2022-09-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Identification and Optimization of a Ligand-Efficient Benzoazepinone Bromodomain and Extra Terminal (BET) Family Acetyl-Lysine Mimetic into the Oral Candidate Quality Molecule I-BET432.
J.Med.Chem., 65, 2022
|
|
