1ZAO
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ATP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
1ZAR
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ADP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
1K2A
 
 | | Modified Form of Eosinophil-derived Neurotoxin | | Descriptor: | SULFATE ION, eosinophil-derived neurotoxin | | Authors: | Chang, C, Newton, D.L, Rybak, S.M, Wlodawer, A. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallographic and functional studies of a modified form of eosinophil-derived neurotoxin (EDN) with novel biological activities.
J.Mol.Biol., 317, 2002
|
|
1K6U
 
 | | Crystal Structure of Cyclic Bovine Pancreatic Trypsin Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Botos, I, Wu, Z, Lu, W, Wlodawer, A. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a cyclic form of bovine pancreatic trypsin inhibitor.
FEBS Lett., 509, 2001
|
|
1L5B
 
 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1L5E
 
 | | The domain-swapped dimer of CV-N in solution | | Descriptor: | Cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1LOM
 
 | | CYANOVIRIN-N DOUBLE MUTANT P51S S52P | | Descriptor: | CALCIUM ION, Cyanovirin-N, SULFATE ION | | Authors: | Botos, I, Mori, T, Cartner, L.K, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Domain-swapped structure of a mutant of cyanovirin-N.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1O7J
 
 | | Atomic resolution structure of Erwinia chrysanthemi L-asparaginase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-ASPARAGINASE, ... | | Authors: | Lubkowski, J, Dauter, M, Aghaiypour, K, Wlodawer, A, Dauter, Z. | | Deposit date: | 2002-11-07 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Erwinia Chrysanthemi L-Asparaginase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7VE0
 
 | | Crystal Structure of Ritonavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
7VE2
 
 | | Crystal Structure of Lopinavir bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, Plasmepsin II | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2021-09-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Plasmodium falciparum plasmepsins by drugs targeting HIV-1 protease: A way forward for antimalarial drug discovery.
Curr Res Struct Biol, 7, 2024
|
|
1RTA
 
 | |
1RTB
 
 | |
7RY7
 
 | |
5YIA
 
 | | Crystal Structure of KNI-10343 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-hydroxyphenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIE
 
 | | Crystal Structure of KNI-10742 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-azanylethyl(ethyl)amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIC
 
 | | Crystal Structure of KNI-10333 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-aminophenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIB
 
 | | Crystal Structure of KNI-10743 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-(dimethylamino)ethyl-methyl-amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YID
 
 | | Crystal Structure of KNI-10395 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
4PGA
 
 | | GLUTAMINASE-ASPARAGINASE FROM PSEUDOMONAS 7A | | Descriptor: | AMMONIUM ION, GLUTAMINASE-ASPARAGINASE, SULFATE ION | | Authors: | Jakob, C.G, Lewinski, K, Lacount, M.W, Roberts, J, Lebioda, L. | | Deposit date: | 1997-01-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ion binding induces closed conformation in Pseudomonas 7A glutaminase-asparaginase (PGA): crystal structure of the PGA-SO4(2-)-NH4+ complex at 1.7 A resolution.
Biochemistry, 36, 1997
|
|
8QEV
 
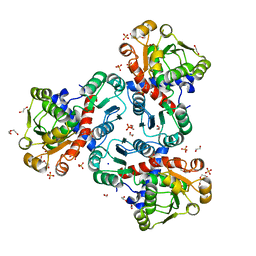 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
8DUF
 
 | |
6X5N
 
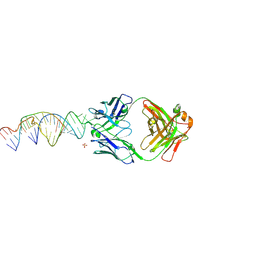 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
3SQF
 
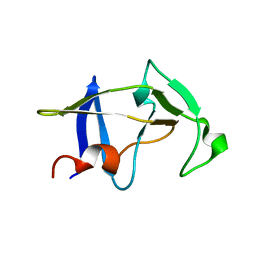 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2WUR
 
 | |
