5T73
 
 | |
6EUD
 
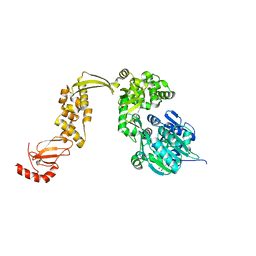 | |
7ACL
 
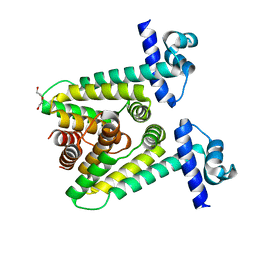 | |
7ACO
 
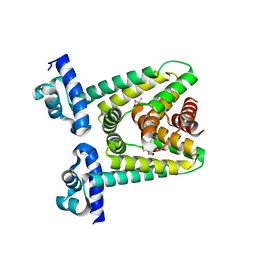 | |
7ACM
 
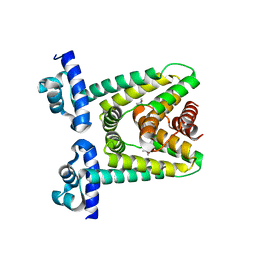 | |
7ZP0
 
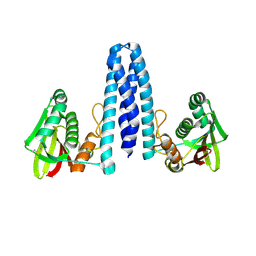 | |
8QEV
 
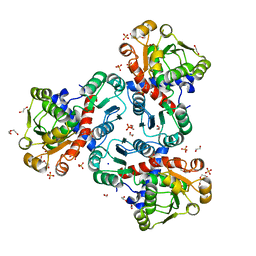 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
5ORF
 
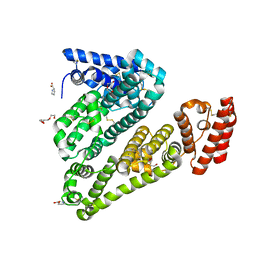 | | Structure of ovine serum albumin in P1 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, PROLINE, Serum albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ORI
 
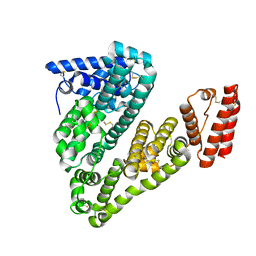 | | Structure of caprine serum albumin in orthorhombic crystal system | | Descriptor: | Albumin | | Authors: | Bujacz, A, Talaj, J.A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6ZVG
 
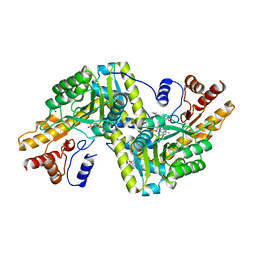 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-indole-3-lactic acid | | Descriptor: | 3-(INDOL-3-YL) LACTATE, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Rum, J, Rutkiewicz, M, Pruska, A, Bujacz, A, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-24 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUP
 
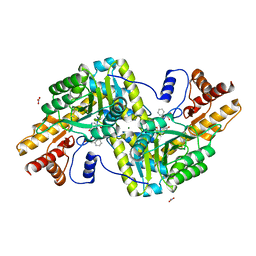 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-(-)-3-phenyllactic acid | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6ZUR
 
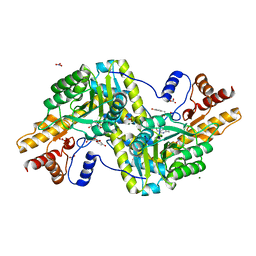 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - L-p-hydroxyphenyllactic acid | | Descriptor: | (2S)-2-hydroxy-3-(4-hydroxyphenyl)propanoic acid, Aminotransferase, MAGNESIUM ION, ... | | Authors: | Bujacz, A, Rum, J, Rutkiewicz, M, Pietrzyk-Brzezinska, A.J, Bujacz, G. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials (Basel), 14, 2021
|
|
6YXQ
 
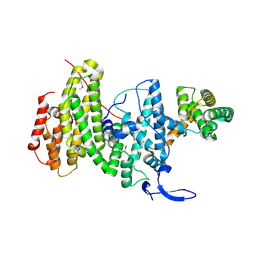 | | Crystal structure of a DNA repair complex ASCC3-ASCC2 | | Descriptor: | Activating signal cointegrator 1 complex subunit 2, Activating signal cointegrator 1 complex subunit 3 | | Authors: | Jia, J, Absmeier, E, Holton, N, Bohnsack, K.E, Pietrzyk-Brzezinska, A.J, Bohnsack, M.T, Wahl, M.C. | | Deposit date: | 2020-05-03 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Nat Commun, 11, 2020
|
|
