5C6W
 
 | | anti-CXCL13 scFv - E10 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimization of a scFv-based biotherapeutic by CDR side-chain clash repair
To Be Published
|
|
3KCF
 
 | |
5CBA
 
 | | 3B4 in complex with CXCL13 - 3B4-CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, 3b4 heavy chain, 3b4 light chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
3BT1
 
 | | Structure of urokinase receptor, urokinase and vitronectin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | Authors: | Huang, M. | | Deposit date: | 2007-12-27 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3BT2
 
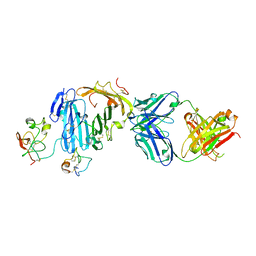 | | Structure of urokinase receptor, urokinase and vitronectin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | Authors: | Huang, M. | | Deposit date: | 2007-12-27 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
3KVX
 
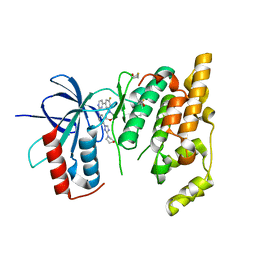 | | JNK3 bound to aminopyrimidine inhibitor, SR-3562 | | Descriptor: | Mitogen-activated protein kinase 10, N-[(2Z)-4-(3-fluoro-5-morpholin-4-ylphenyl)pyrimidin-2(1H)-ylidene]-4-(3-morpholin-4-yl-1H-1,2,4-triazol-1-yl)aniline | | Authors: | Habel, J.E, Laughlin, J.D, LoGrasso, P. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors
J.Med.Chem., 53, 2010
|
|
4NTG
 
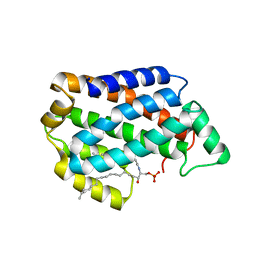 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
2XR4
 
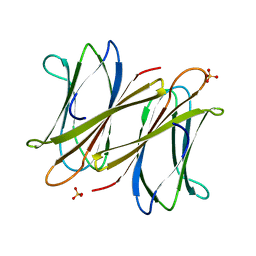 | | C-terminal domain of BC2L-C Lectin from Burkholderia cenocepacia | | Descriptor: | LECTIN, SULFATE ION | | Authors: | Sulak, O, Cioci, G, Lameignere, E, Delia, M, Wimmerova, M, Imberty, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Burkholderia Cenocepacia Bc2L-C is a Super Lectin with Dual Specificity and Proinflammatory Activity.
Plos Pathog., 7, 2011
|
|
3HNB
 
 | |
3HOB
 
 | |
3HNY
 
 | |
8RIH
 
 | |
1XJD
 
 | |
6E7I
 
 | | Human ppGalNAcT2 I253A/L310A Mutant with EA2 and UDP | | Descriptor: | EA2, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, ... | | Authors: | Bertozzi, C.R, Schumann, B, Agbay, A.J. | | Deposit date: | 2018-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
6NQT
 
 | | GalNac-T2 soaked with UDP-sugar | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{R})-3-(hex-5-ynoylamino)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Fernandez, D, Bertozzi, C.R, Schumann, B, Agbay, A. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
1RSM
 
 | | THE 2-ANGSTROMS RESOLUTION STRUCTURE OF A THERMOSTABLE RIBONUCLEASE A CHEMICALLY CROSS-LINKED BETWEEN LYSINE RESIDUES 7 AND 41 | | Descriptor: | DINITROPHENYLENE, RIBONUCLEASE A | | Authors: | Weber, P.C, Sheriff, S, Ohlendorf, D.H, Finzel, B.C, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2-A resolution structure of a thermostable ribonuclease A chemically cross-linked between lysine residues 7 and 41.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
7QL7
 
 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 9 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
6L08
 
 | |
7N18
 
 | | Clostridium botulinum Neurotoxin Serotype A Light Chain Inhibited by a Chiral Hydroxamic Acid | | Descriptor: | (3R)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, (3S)-3-(4-chlorophenyl)-N,5-dihydroxypentanamide, Botulinum neurotoxin type A, ... | | Authors: | Silvaggi, N.R, Allen, K.N. | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Use of Crystallography and Molecular Modeling for the Inhibition of the Botulinum Neurotoxin A Protease.
Acs Med.Chem.Lett., 12, 2021
|
|
3K45
 
 | | Alternate Binding Modes Observed for the E- and Z-isomers of 2,4-Diaminofuro[2,3d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3K47
 
 | | Alternate Binding Modes Observed for the E- and Z-Isomers of 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Queener, S.F, Gangjee, A. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
4FSF
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3FXY
 
 | | Acidic Mammalian Chinase, Catalytic Domain | | Descriptor: | Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3FY1
 
 | | The Acidic Mammalian Chitinase catalytic domain in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
