6PWW
 
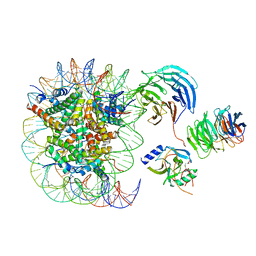 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
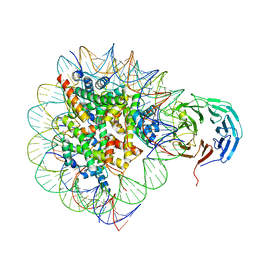 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
9ERJ
 
 | |
9ERL
 
 | |
7LAW
 
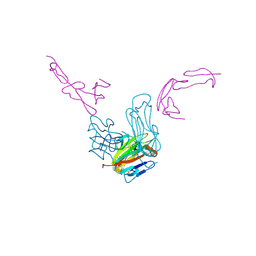 | | crystal structure of GITR complex with GITR-L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Longenecker, K.L, Rogers, B, Bigelow, L, Judge, R.A, Alvarez, H. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | An anti-PD-1-GITR-L bispecific agonist induces GITR clustering-mediated T cell activation for cancer immunotherapy.
Nat Cancer, 3, 2022
|
|
9HCJ
 
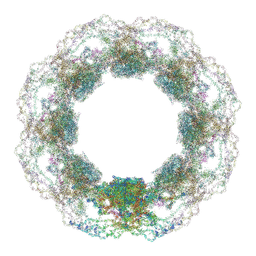 | | D. discoideum nuclear pore complex | | Descriptor: | ELYS-like domain-containing protein, NDC1, Nuclear pore complex protein, ... | | Authors: | Obarska-Kosinska, A, Hoffmann, P.C, Beck, M. | | Deposit date: | 2024-11-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Nuclear pore permeability and fluid flow are modulated by its dilation state.
Mol.Cell, 85, 2025
|
|
9ERK
 
 | |
9ERI
 
 | |
4QBS
 
 | |
6XJF
 
 | |
4QBR
 
 | |
1LCP
 
 | |
8HFB
 
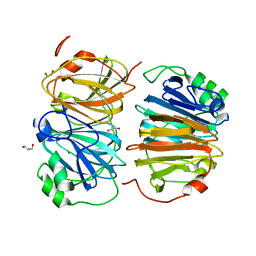 | | Evolved variant of quercetin 2,4-dioxygenase from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Eom, H, Song, W.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Underlying Role of Hydrophobic Environments in Tuning Metal Elements for Efficient Enzyme Catalysis.
J.Am.Chem.Soc., 145, 2023
|
|
6JQ7
 
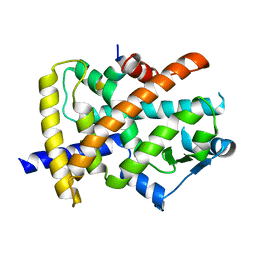 | |
6XDM
 
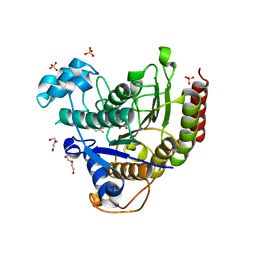 | |
6IJR
 
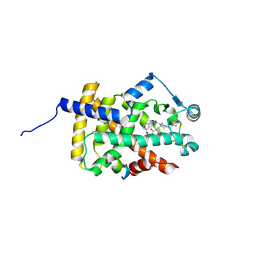 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6IJS
 
 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
9BDQ
 
 | | The structure of NiV L-P complex | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Hu, S, Yang, P, Yu, Z, Abraham, J. | | Deposit date: | 2024-04-12 | | Release date: | 2025-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structural and functional analysis of the Nipah virus polymerase complex.
Cell, 188, 2025
|
|
3KPE
 
 | | Solution structure of the respiratory syncytial virus (RSV)six-helix bundle complexed with TMC353121, a small-moleucule inhibitor of RSV | | Descriptor: | 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, Fusion glycoprotein F0, TETRAETHYLENE GLYCOL | | Authors: | Roymans, D, De Bondt, H, Arnoult, E, Cummings, M.D, Van Vlijmen, H, Andries, K. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5YI5
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7M72
 
 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxy-15-methylhexadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
7CM0
 
 | | Crystal structure of a glutaminyl cyclase in complex with NHV-1009 | | Descriptor: | 1-(cyclopentylmethyl)-1-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]-3-[3-(5-methylimidazol-1-yl)propyl]urea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Lee, J.W, Song, J.Y, Jang, T.H, Ha, J.H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of highly potent human glutaminyl cyclase (QC) inhibitors as anti-Alzheimer's agents by the combination of pharmacophore-based and structure-based design.
Eur.J.Med.Chem., 226, 2021
|
|
2K21
 
 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2LYQ
 
 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
