3ITK
 
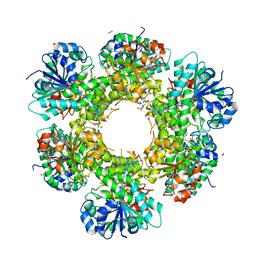 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
7AKG
 
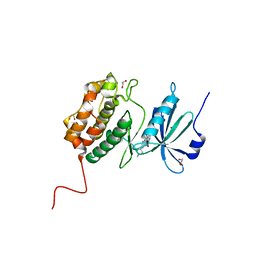 | |
7P7F
 
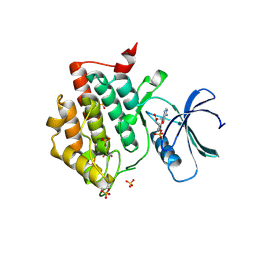 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7P7G
 
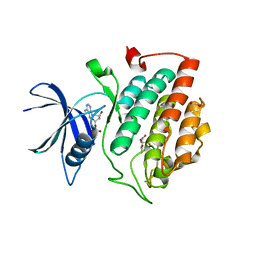 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
7P7H
 
 | |
7APF
 
 | | Crystal structure of JAK3 in complex with FM601 (compound 10a) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 3-[3-(propanoylamino)phenyl]-1~{H}-pyrrolo[2,3-b]pyridine-5-carboxamide, ... | | Authors: | Chaikuad, A, Forster, M, Gehringer, M, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a Novel Class of Covalent Dual Inhibitors Targeting the Protein Kinases BMX and BTK.
Int J Mol Sci, 21, 2020
|
|
7AYI
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2a) | | Descriptor: | 7-(2-phenylazanylpyrimidin-4-yl)-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
7APG
 
 | | Crystal structure of JAK3 in complex with FM587 (compound 9a) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, Tyrosine-protein kinase JAK3, ... | | Authors: | Chaikuad, A, Forster, M, Gehringer, M, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Covalent Dual Inhibitors Targeting the Protein Kinases BMX and BTK.
Int J Mol Sci, 21, 2020
|
|
7AWC
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
7AYH
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
7B88
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with S99 inhibitor | | Descriptor: | 3-[5-[3,5-bis(chloranyl)phenyl]-4-phenyl-1,3-oxazol-2-yl]propanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Schierle, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Oxaprozin Analogues as Selective RXR Agonists with Superior Properties and Pharmacokinetics.
J.Med.Chem., 64, 2021
|
|
7AWD
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with garcinoic acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
7B10
 
 | | Crystal structure of MLLT1 YEATS domain T1 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, IODIDE ION, ... | | Authors: | Chaikuad, A, Ni, X, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
4URJ
 
 | | Crystal structure of human BJ-TSA-9 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN FAM83A | | Authors: | Pinkas, D.M, Sanvitale, C, Wang, D, Krojer, T, Kopec, J, Chaikuad, A, Dixon Clarke, S, Berridge, G, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Bj-Tsa-9
To be Published
|
|
4UUC
 
 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
4UYI
 
 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
3S95
 
 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
6TBE
 
 | | LC3A in complex with (3R,4S,5R,6R)-5-hydroxy-6-((4-hydroxy-3-(4-hydroxy-3-isopentylbenzamido)-8-methyl-2-oxo-2H-chromen-7-yl)oxy)-3-methoxy-2,2-dimethyltetrahydro-2H-pyran-4-yl carbamate | | Descriptor: | 1,2-ETHANEDIOL, Microtubule-associated proteins 1A/1B light chain 3A, NOVOBIOCIN | | Authors: | Kramer, J.S, Pogoryelov, D, Hartmann, M, Chaikuad, A, Proschak, E. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67008042 Å) | | Cite: | Demonstrating Ligandability of the LC3A and LC3B Adapter Interface.
J.Med.Chem., 64, 2021
|
|
7BF4
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with GMP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, NSP3 macrodomain | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF3
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, MAGNESIUM ION, ... | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7BF6
 
 | | Crystal structure of SARS-CoV-2 macrodomain in complex with remdesivir metabolite GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Ni, X, Knapp, S, Chaikuad, A, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-31 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
7B9O
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with S169 inhibitor | | Descriptor: | 3-(5-(3,5-bis(trifluoromethyl)phenyl)-4-phenyloxazol-2-yl)propanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Ni, X, Chaikuad, A, Schierle, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Oxaprozin Analogues as Selective RXR Agonists with Superior Properties and Pharmacokinetics.
J.Med.Chem., 64, 2021
|
|
4O38
 
 | | Crystal structure of the human cyclin G associated kinase (GAK) | | Descriptor: | Cyclin-G-associated kinase, GLYCEROL, SUCCINIC ACID | | Authors: | Zhang, R, Hatzos-Skintges, C, Weger, A, Chaikuad, A, Eswaran, J, Fedorov, O, King, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the human cyclin G associated kinase (GAK)
To be Published
|
|
7Z7E
 
 | | Crystal structure of p63 DNA binding domain in complex with inhibitory DARPin G4 | | Descriptor: | DARPIN, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Strubel, A, Gebel, J, Chaikuad, A, Muenick, P, Doetsch, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
5MWA
 
 | | human sEH Phosphatase in complex with 3-4-3,4-dichlorophenyl-5-phenyl-1,3-oxazol-2-yl-benzoic-acid | | Descriptor: | 3-[4-(3,4-dichlorophenyl)-5-phenyl-1,3-oxazol-2-yl]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Sorrell, F.J, Fox, N, Chaikuad, A, Knapp, S, Proschak, E. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of first in vivo active inhibitors of soluble epoxide hydrolase (sEH) phosphatase domain.
J.Med.Chem., 2019
|
|
