6DTT
 
 | | Apo T. maritima MalE2 | | Descriptor: | maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S, Myles, D.A. | | Deposit date: | 2022-06-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTQ
 
 | | Maltose bound T. maritima MalE3 | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTR
 
 | | Apo T. maritima MalE3 | | Descriptor: | SULFATE ION, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
9DSY
 
 | | Crystal Structure of C4-Dicarboxylate-Binding Periplasmic Protein (PA5167) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid | | Descriptor: | C4-dicarboxylate-binding periplasmic protein DctP, SUCCINIC ACID, ZINC ION | | Authors: | Minasov, G, Shukla, S, Shuvalova, L, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of C4-Dicarboxylate-Binding Periplasmic Protein (PA5167) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid
To Be Published
|
|
9DTL
 
 | | Crystal Structure of C4-Dicarboxylate-Binding Protein (PA0884) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid | | Descriptor: | C4-dicarboxylate-binding periplasmic protein, GLYCEROL, SUCCINIC ACID | | Authors: | Minasov, G, Shukla, S, Shuvalova, L, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-10-01 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of C4-Dicarboxylate-Binding Protein (PA0884) of Tripartite ATP-independent Periplasmic Transporter Family from Pseudomonas aeruginosa PAO1 in Complex with Succinic Acid
To Be Published
|
|
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
7ND1
 
 | | First-in-class small molecule inhibitors of Polycomb Repressive Complex 1 (PRC1) RING domain | | Descriptor: | 3-(2-chlorophenyl)-4-ethyl-5-(1~{H}-indol-4-yl)-1~{H}-pyrrole-2-carboxylic acid, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, ... | | Authors: | Cierpicki, T, Lund, G, Jaremko, L. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI7
 
 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI8
 
 | |
4W9Y
 
 | |
6KQQ
 
 | | NSD1 SET domain in complex with BT3 and SAM | | Descriptor: | 2-azanyl-6-[(2-azanyl-4-oxidanyl-1,3-benzothiazol-6-yl)disulfanyl]-1,3-benzothiazol-4-ol, 2-azanyl-6-sulfanyl-1,3-benzothiazol-4-ol, CALCIUM ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6KQP
 
 | | NSD1 SET domain in complex with SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
3MH8
 
 | |
3MH9
 
 | |
3MHA
 
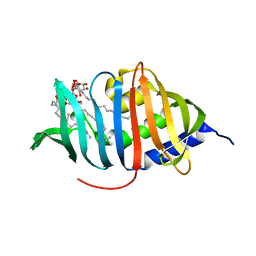 | | Crystal structure of LprG from Mycobacterium tuberculosis bound to PIM | | Descriptor: | (1S,2R,3R,4S,5R,6S)-2-(alpha-L-allopyranosyloxy)-3,4,5-trihydroxy-6-({6-O-[(1R)-1-hydroxyhexadecyl]-beta-L-gulopyranosyl}oxy)cyclohexyl (2R)-2-{[(1S)-1-hydroxyhexadecyl]oxy}-3-{[(1S,10S)-1-hydroxy-10-methyloctadecyl]oxy}propyl hydrogen (R)-phosphate, Lipoprotein lprG | | Authors: | Tsai, H.-C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-07 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5JUE
 
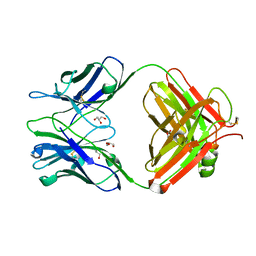 | | Crystal Structure of UIC2 Fab | | Descriptor: | GLYCEROL, heavy chain of UIC2 Fab, light chain of UIC2 Fab | | Authors: | Xia, D, Esser, L. | | Deposit date: | 2016-05-10 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the antigen-binding fragment of a monoclonal antibody specific for the multidrug-resistance-linked ABC transporter human P-glycoprotein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5KO2
 
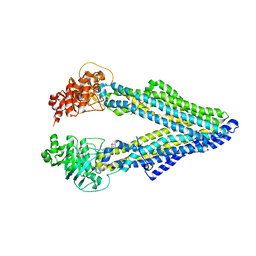 | | Mouse pgp 34 linker deleted mutant Hg derivative | | Descriptor: | MERCURY (II) ION, Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
5KOY
 
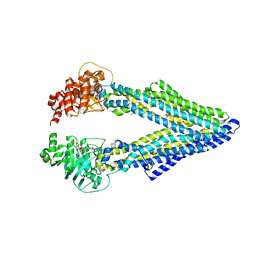 | | Mouse pgp 34 linker deleted bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Multidrug resistance protein 1A | | Authors: | Xia, D, Esser, L, Zhou, F. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity.
J. Biol. Chem., 292, 2017
|
|
