6IXD
 
 | | X-ray crystal structure of bPI-11 hiv-1 protease complex | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Adachi, M, Hidaka, K. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Acquired Removability of Aspartic Protease Inhibitors by Direct Biotinylation.
Bioconjug.Chem., 30, 2019
|
|
5YOJ
 
 | | Structure of A17 HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, A17 HIV-1 protease, GLYCEROL | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
5YOK
 
 | | Structure of HIV-1 Protease in Complex with Inhibitor KNI-1657 | | Descriptor: | (4R)-N-[(2,6-dimethylphenyl)methyl]-3-[(2S,3S)-3-[[(2S)-2-[(7-methoxy-1-benzofuran-2-yl)carbonylamino]-2-[(3R)-oxolan-3 -yl]ethanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, HIV-1 PROTEASE | | Authors: | Adachi, M, Hidaka, K, Kuroki, R, Kiso, Y. | | Deposit date: | 2017-10-29 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Identification of Highly Potent Human Immunodeficiency Virus Type-1 Protease Inhibitors against Lopinavir and Darunavir Resistant Viruses from Allophenylnorstatine-Based Peptidomimetics with P2 Tetrahydrofuranylglycine.
J. Med. Chem., 61, 2018
|
|
3A2O
 
 | | Crystal Structure of HIV-1 Protease Complexed with KNI-1689 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(4-amino-2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-5,5-dimethyl-N-(2-methylprop -2-en-1-yl)-1,3-thiazolidine-4-carboxamide, GLYCEROL, PROTEASE | | Authors: | Adachi, M, Tamada, T, Hidaka, K, Kimura, T, Kiso, Y, Kuroki, R. | | Deposit date: | 2009-05-26 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2' pocket.
J.Med.Chem., 52, 2009
|
|
2ANL
 
 | | X-ray crystal structure of the aspartic protease plasmepsin 4 from the malarial parasite plasmodium malariae bound to an allophenylnorstatine based inhibitor | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, plasmepsin IV | | Authors: | Clemente, J.C, Govindasamy, L, Madabushi, A, Fisher, S.Z, Moose, R.E, Yowell, C.A, Hidaka, K, Kimura, T, Hayashi, Y, Kiso, Y, Agbandje-McKenna, M, Dame, J.B, Dunn, B.M, McKenna, R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8WU5
 
 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
3FX5
 
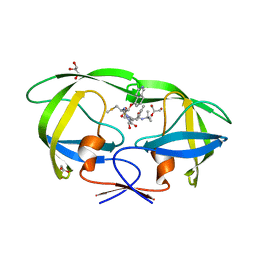 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDC
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10074 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dichlorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
3KDD
 
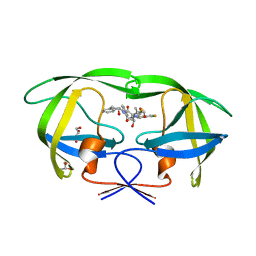 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
5UGG
 
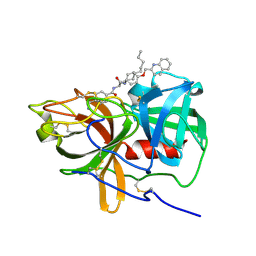 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(quinolin-2-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
5UGD
 
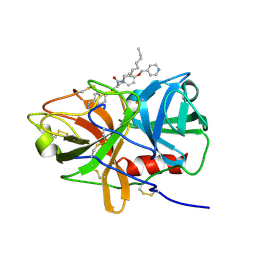 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(pyridin-4-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
7E6P
 
 | | Fab-amyloid beta fragment complex | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta fragment with an intramolecular disulfide bond at positions 17 and 28, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Miki, K. | | Deposit date: | 2021-02-23 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of a Conformation-Restricted Amyloid beta Peptide and Immunoreactivity of Its Antibody in Human AD brain.
Acs Chem Neurosci, 12, 2021
|
|
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3QS1
 
 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
3QRV
 
 | |
3QVC
 
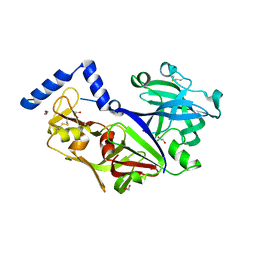 | |
5YIA
 
 | | Crystal Structure of KNI-10343 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-hydroxyphenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIE
 
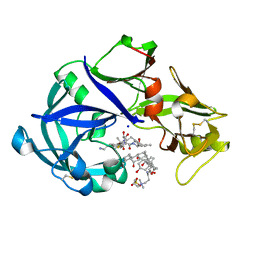 | | Crystal Structure of KNI-10742 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-azanylethyl(ethyl)amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIC
 
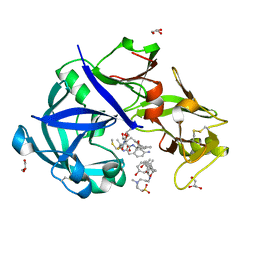 | | Crystal Structure of KNI-10333 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2R)-2-[2-(4-aminophenyl)ethanoylamino]-3-methylsulfanyl-propanoyl]amino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, GLYCEROL, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YIB
 
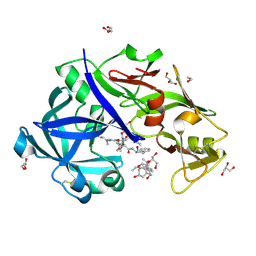 | | Crystal Structure of KNI-10743 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-(dimethylamino)ethyl-methyl-amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
5YID
 
 | | Crystal Structure of KNI-10395 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]ami no}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
