1MAB
 
 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1K12
 
 | | Fucose Binding lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, LECTIN, ... | | Authors: | Bianchet, M.A, Odom, E.W, Vasta, G.R, Amzel, L.M. | | Deposit date: | 2001-09-23 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel fucose recognition fold involved in innate immunity.
Nat.Struct.Biol., 9, 2002
|
|
3CQO
 
 | |
1Q3F
 
 | | Uracil DNA glycosylase bound to a cationic 1-aza-2'-deoxyribose-containing DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*(NRI)P*AP*TP*CP*TP*T)-3', PHOSPHATE ION, ... | | Authors: | Bianchet, M.A, Seiple, L.A, Jiang, Y.L, Ichikawa, Y, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2003-07-29 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase.
Biochemistry, 42, 2003
|
|
5DC2
 
 | | X-RAY CRYSTAL STRUCTURE OF A ENZYMATICALLY DEGRADED BIAPENEM-ADDUCT OF L,D-TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5DCC
 
 | | X-RAY CRYSTAL STRUCTURE OF a TEBIPENEM ADDUCT OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
1A78
 
 | | COMPLEX OF TOAD OVARY GALECTIN WITH THIO-DIGALACTOSE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GALECTIN-1, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1998-03-20 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes
Proteins, 40, 2000
|
|
3FCK
 
 | | Complex of UNG2 and a fragment-based design inhibitor | | Descriptor: | 3-({[3-({[(1E)-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methylidene]amino}oxy)propyl]amino}methyl)benzoic acid, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCI
 
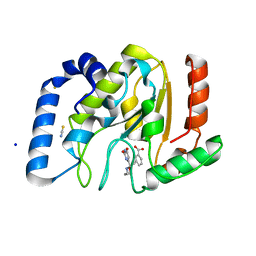 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{(E)-[(3-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}propoxy)imino]methyl}benzoic acid, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCF
 
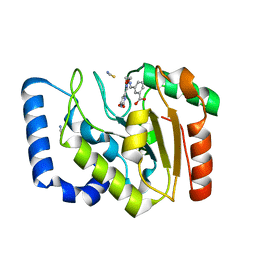 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-[(1E,7E)-8-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)-3,6-dioxa-2,7-diazaocta-1,7-dien-1-yl]benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3FCL
 
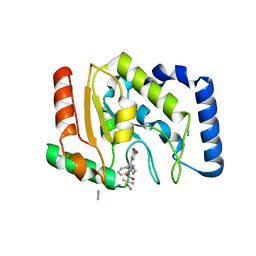 | | Complex of UNG2 and a fragment-based designed inhibitor | | Descriptor: | 3-{[(4-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}butyl)amino]methyl}benzoic acid, THIOCYANATE ION, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2008-11-21 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
6E0T
 
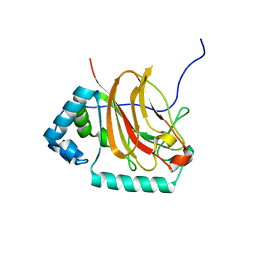 | | C-terminal domain of Fission Yeast OFD1 | | Descriptor: | Prolyl 3,4-dihydroxylase ofd1 | | Authors: | Bianchet, M.A, Amzel, L.M, Espenshade, P.J, Yeh, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The hypoxic regulator of sterol synthesis nro1 is a nuclear import adaptor.
Structure, 19, 2011
|
|
5D7H
 
 | | X-RAY CRYSTAL STRUCTURE OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYCEROL, L,D-transpeptidase 2, SULFATE ION | | Authors: | Saavedra, H, Basta, L.A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
3TUR
 
 | | Crystal Structure of M. tuberculosis LD-transpeptidase type 2 complexed with a peptidoglycan fragment | | Descriptor: | 6-CARBOXYLYSINE, D-GLUTAMIC ACID, Di-mu-iodobis(ethylenediamine)diplatinum(II), ... | | Authors: | Bianchet, M.A, Erdemli, S.B, Gupta, R, Lamichhane, G, Amzel, L.M. | | Deposit date: | 2011-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Targeting the Cell Wall of Mycobacterium tuberculosis: Structure and Mechanism of L,D-Transpeptidase 2.
Structure, 20, 2012
|
|
2HXM
 
 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2OXM
 
 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2OYT
 
 | | Crystal Structure of UNG2/DNA(TM) | | Descriptor: | DNA strand1, DNA strand2, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
1PBO
 
 | | COMPLEX OF BOVINE ODORANT BINDING PROTEIN (OBP) WITH A SELENIUM CONTAINING ODORANT | | Descriptor: | 4-butyl-5-propyl-1,3-selenazol-2-amine, ODORANT BINDING PROTEIN | | Authors: | Amzel, L.M, Bianchet, M.A, Monaco, H, Bains, G. | | Deposit date: | 1996-07-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of bovine odorant binding protein and its mechanism of odor recognition.
Nat.Struct.Biol., 3, 1996
|
|
1GAN
 
 | | COMPLEX OF TOAD OVARY GALECTIN WITH N-ACETYLGALACTOSE | | Descriptor: | GALECTIN-1, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Amzel, L.M, Bianchet, M.A, Ahmed, H, Vasta, G.R. | | Deposit date: | 1996-11-06 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes.
Proteins, 40, 2000
|
|
8GB1
 
 | | Crystal structure of SAMHD1 dimer bound to deoxyguanosine linked inhibitor | | Descriptor: | 5'-O-[(R)-(3-{[(1M)-3'-bromo[1,1'-biphenyl]-3-carbonyl]amino}propoxy)(hydroxy)phosphoryl]-2'-deoxyguanosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
8GB2
 
 | | Crystal structure of Apo-SAMHD1 | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Deoxyguanosine-Linked Bifunctional Inhibitor of SAMHD1 dNTPase Activity and Nucleic Acid Binding.
Acs Chem.Biol., 18, 2023
|
|
1QR2
 
 | | HUMAN QUINONE REDUCTASE TYPE 2 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (QUINONE REDUCTASE TYPE 2), ZINC ION | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-15 | | Release date: | 1999-08-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
1QRD
 
 | | QUINONE REDUCTASE/FAD/CIBACRON BLUE/DUROQUINONE COMPLEX | | Descriptor: | CIBACRON BLUE, DUROQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, R, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1995-07-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
6BOI
 
 | | Crystal Structure of LdtMt2 (56-408) with a panipenem adduct at the active site cysteine-354 | | Descriptor: | (3S,5R)-5-[(2R,3R)-1,3-dihydroxybutan-2-yl]-3-({(3R)-1-[(1E)-ethanimidoyl]pyrrolidin-3-yl}sulfanyl)-L-proline, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Saavedra, H, Bianchet, M.A. | | Deposit date: | 2017-11-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures and Mechanism of Inhibition of Mycobacterium tuberculosis L,D-transpeptidase 2 by Panipenem
To Be Published
|
|
6E20
 
 | | Crystal structure of the Dario rerio galectin-1-L2 | | Descriptor: | Galectin, MAGNESIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ghosh, A, Bianchet, M.A. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Glycobiology, 29, 2019
|
|
