2XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
4YWQ
 
 | | Crystal structure of the ROQ domain of human Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-20 | | Release date: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ROQ domain of human Roquin-1
To be Published
|
|
2XJL
 
 | | Monomeric Human Cu,Zn Superoxide dismutase without Cu ligands | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Saraboji, K, Leinartaite, L, Nordlund, A, Oliveberg, M, Logan, D.T. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Folding Catalysis by Transient Coordination of Zn2+ to the Cu Ligands of the Als-Associated Enzyme Cu/Zn Superoxide Dismutase 1.
J.Am.Chem.Soc., 132, 2010
|
|
4YM5
 
 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.277 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
4YMY
 
 | | Crystal structure of mutant nitrobindin M75A/H76L/Q96C/M148L/H158A (NB11) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Himiyama, T, Tachikawa, K, Oohora, K, Onoda, A, Hayashi, T. | | Deposit date: | 2015-03-08 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A Highly Active Biohybrid Catalyst for Olefin Metathesis in Water: Impact of a Hydrophobic Cavity in a beta-Barrel Protein
Acs Catalysis, 5, 2015
|
|
4YP3
 
 | | Mutant Human DNA Polymerase Eta Q38A/R61A Inserting dCTP Opposite an 8-Oxoguanine Lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Su, Y, Patra, A, Harp, J.M, Egli, M, Guengerich, F.P. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Roles of Residues Arg-61 and Gln-38 of Human DNA Polymerase eta in Bypass of Deoxyguanosine and 7,8-Dihydro-8-oxo-2'-deoxyguanosine.
J.Biol.Chem., 290, 2015
|
|
6BIT
 
 | | SIRPalpha antibody complex | | Descriptor: | KWAR23 Fab heavy chain, KWAR23 Fab light chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Ring, N.G, Herndler-Brandstetter, D, Weiskopf, K, Shan, L, Volkmer, J.P, George, B.M, Lietzenmayer, M, McKenna, K.M, Naik, T.J, McCarty, A, Zheng, Y, Ring, A.M, Flavell, R.A, Weissman, I.L. | | Deposit date: | 2017-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Anti-SIRP alpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YO5
 
 | | EAEC T6SS TssA-Cterminus | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
2XGL
 
 | | The X-ray structure of the Escherichia coli colicin M immunity protein demonstrates the presence of a disulphide bridge, which is functionally essential | | Descriptor: | CADMIUM ION, CHLORIDE ION, COLICIN-M IMMUNITY PROTEIN, ... | | Authors: | Gerard, F, Brooks, M.A, Barreteau, H, Touze, T, Graille, M, Bouhss, A, Blanot, D, Tilbeurgh, H.v, Mengin-Lecreulx, D. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-17 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray Structure and Site-Directed Mutagenesis Analysis of the Escherichia Coli Colicin M Immunity Protein.
J.Bacteriol., 193, 2011
|
|
2XBI
 
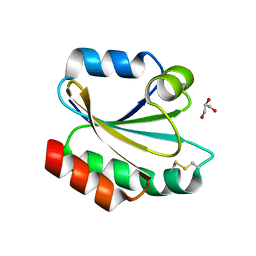 | | Crystal structure of Schistosoma mansoni Thioredoxin at 1.6 Angstrom | | Descriptor: | GLYCEROL, THIOREDOXIN | | Authors: | Boumis, G, Miele, A.E, Dimastrogiovanni, D, Angelucci, F, Bellelli, A. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of Schistosoma Mansoni Thioredoxin.
Protein Sci., 20, 2011
|
|
6D2P
 
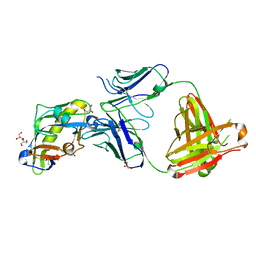 | |
3M8U
 
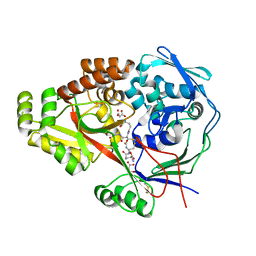 | | Crystal structure of glutathione-binding protein A (GbpA) from Haemophilus parasuis SH0165 in complex with glutathione disulfide (GSSG) | | Descriptor: | Heme-binding protein A, MALONATE ION, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Vergauwen, B, Elegheert, J, Dansercoer, A, Devreese, B, Savvides, S.N. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6IVX
 
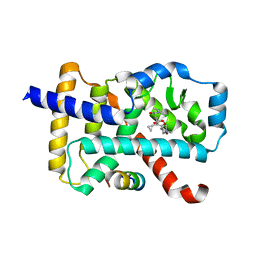 | | Discovery of the Second Generation ROR gamma Inhibitors Composed of an Azole Scaffold. | | Descriptor: | (4S)-4-[4'-cyclopropyl-5-(2,2-dimethylpropyl)[3,5'-bi-1,2-oxazol]-3'-yl]-6-[(2,4-dichlorophenyl)amino]-6-oxohexanoic acid, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Adachi, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Second Generation ROR gamma Inhibitors Composed of an Azole Scaffold.
J. Med. Chem., 62, 2019
|
|
6CDH
 
 | | Crystal structure of ferrous form of the Cl-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
4YT6
 
 | |
4YTB
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
3QTB
 
 | | Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, Uncharacterized protein | | Authors: | Tkaczuk, K.L, Shumilin, I.A, Chruszcz, M, Cymborowski, M, Xu, X, Di Leo, R, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
6D0X
 
 | | Crystal structure of chimeric H.2140 / K.1210 Fab in complex with circumsporozoite protein NANP3 | | Descriptor: | 1210 Antibody, light chain, 2140 Antibody, ... | | Authors: | Scally, S.W, Bosch, A, Imkeller, K, Wardemann, H, Julien, J.P. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-13 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Science, 360, 2018
|
|
6D1M
 
 | | Design, synthesis, and X-ray of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects | | Descriptor: | 4-(cyclohexylselanyl)benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, di Cesare Mannelli, L, Micheli, L, Ghelardini, C, Supuran, C.T. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, synthesis and X-ray crystallography of selenides bearing benzenesulfonamide moiety with neuropathic pain modulating effects.
Eur J Med Chem, 154, 2018
|
|
1UHS
 
 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
6IGK
 
 | | Crystal Structure of human ETB receptor in complex with Endothelin-3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, Kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
6CGO
 
 | | Molecular basis for condensation domain-mediated chain release from the enacyloxin polyketide synthase | | Descriptor: | Condensation domain protein, PHOSPHATE ION | | Authors: | Valentic, T.R, Tsai, S.C, Challis, G.L, Lewandowski, J.R, Kosol, S, Gallo, A, Griffiths, D, Masschelein, J.L, Jenner, M, De los Santos, E. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase.
Nat.Chem., 11, 2019
|
|
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
