6RMH
 
 | | The Rigid-body refined model of the normal Huntingtin. | | Descriptor: | Huntingtin | | Authors: | Jung, T, Tamo, G, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2019-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
6AGO
 
 | | Crystal structure of MRG15-ASH1L Histone methyltransferase complex | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, Mortality factor 4 like 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y, Song, J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis of MRG15-Mediated Activation of the ASH1L Histone Methyltransferase by Releasing an Autoinhibitory Loop.
Structure, 27, 2019
|
|
6YEJ
 
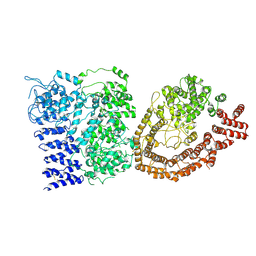 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
6WW5
 
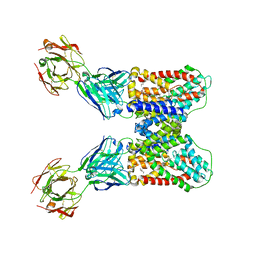 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
4HVZ
 
 | |
4K6J
 
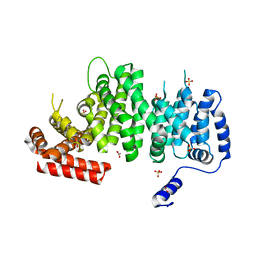 | | Human cohesin inhibitor WapL | | Descriptor: | ACETATE ION, SULFATE ION, Wings apart-like protein homolog | | Authors: | Tomchick, D.R, Yu, H, Ouyang, Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6205 Å) | | Cite: | Structure of the human cohesin inhibitor Wapl.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5JHL
 
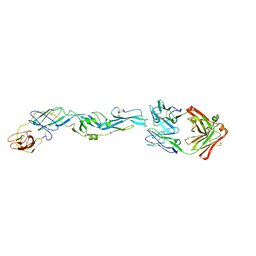 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
5JHM
 
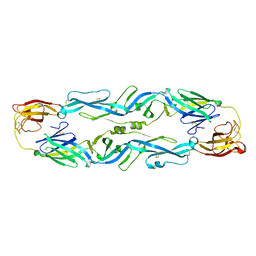 | |
8ESM
 
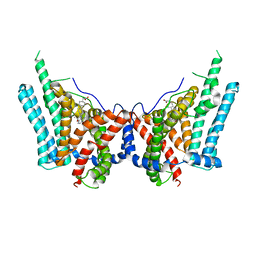 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ETM
 
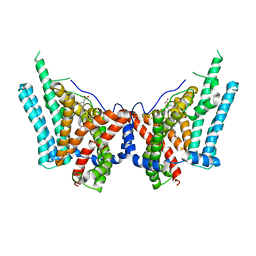 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
6WU1
 
 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU2
 
 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WTW
 
 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU3
 
 | |
6WU4
 
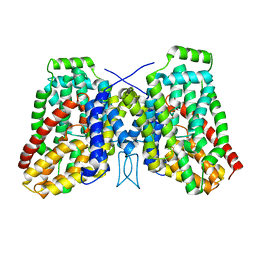 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
2QCY
 
 | |
3CKH
 
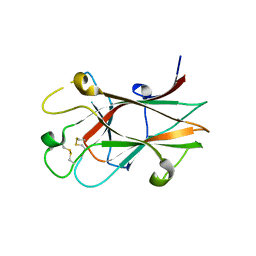 | | Crystal structure of Eph A4 receptor | | Descriptor: | Ephrin type-A receptor 4 | | Authors: | Shi, J.H, Song, J.X. | | Deposit date: | 2008-03-15 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure and NMR Binding Reveal That Two Small Molecule Antagonists Target the High Affinity Ephrin-binding Channel of the EphA4 Receptor.
J.Biol.Chem., 283, 2008
|
|
2BF3
 
 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing ten N-terminal residues (delta-N10 T4moD) | | Descriptor: | HEPTANE-1,2,3-TRIOL, TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2BF5
 
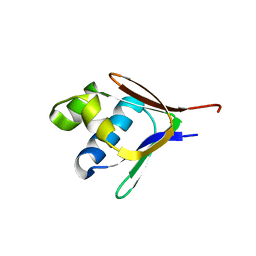 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing four N-terminal residues (delta-N4 T4moD) | | Descriptor: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
7LO7
 
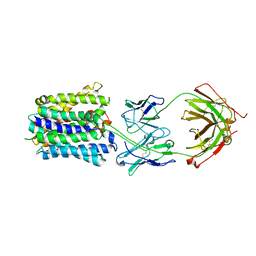 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO8
 
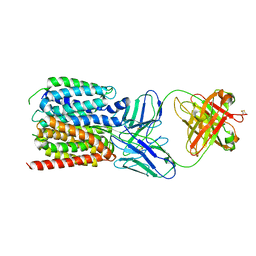 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
6JNF
 
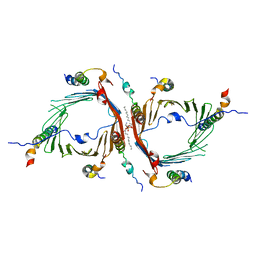 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
3FHQ
 
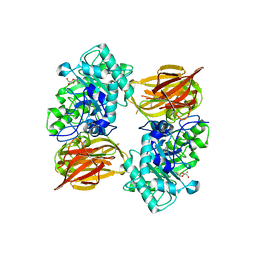 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
3SXN
 
 | | Mycobacterium tuberculosis Eis protein initiates modulation of host immune responses by acetylation of DUSP16/MKP-7 | | Descriptor: | COENZYME A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J.S, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7T9F
 
 | | Structure of VcINDY-apo | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Song, J.M, Wang, D.N. | | Deposit date: | 2021-12-19 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of ion - substrate coupling in the Na + -dependent dicarboxylate transporter VcINDY.
Nat Commun, 13, 2022
|
|
