7NIV
 
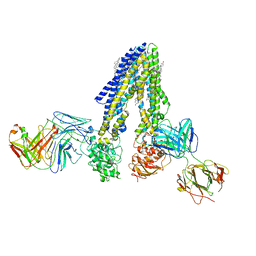 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NIW
 
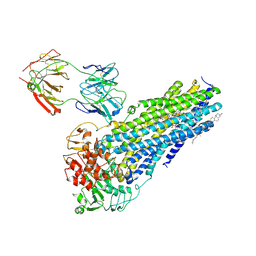 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VKD
 
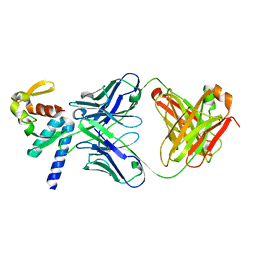 | | Crystal structure of C-terminal domain of Ebola (Bundibugyo) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab light chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A, Derewenda, Z.S. | | Deposit date: | 2017-04-21 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7PGB
 
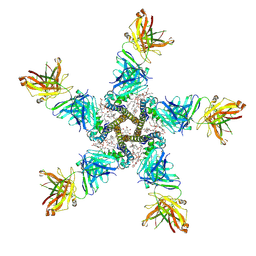 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PG8
 
 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
 | |
7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8VC7
 
 | | Crystal Structure of Human BTN2A1 ectodomain in complex with Antagonist 2A1.9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, Human IgG1 Fragment Antibody Heavy Chain, ... | | Authors: | Ramesh, A, Roy, S, Adams, E. | | Deposit date: | 2023-12-13 | | Release date: | 2024-12-18 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Mapping the extracellular molecular architecture of the pAg-signaling complex with alpha-Butyrophilin antibodies.
Sci Rep, 15, 2025
|
|
8VJ9
 
 | |
6VI2
 
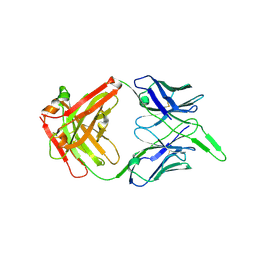 | | Structure of the unaligned Fab4 | | Descriptor: | FAB4 heavy chain, FAB4 light chain | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8VZN
 
 | |
8VZO
 
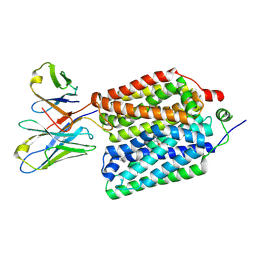 | |
8VTI
 
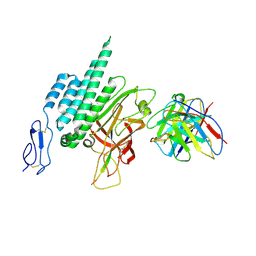 | | Latrophilin-3 (ADGRL3) HormR and GAIN domains in the context of the holoreceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Adhesion G protein-coupled receptor L3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kordon, S.P, Bandekar, S.J, Arac, D. | | Deposit date: | 2024-01-26 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational coupling between extracellular and transmembrane domains modulates holo-adhesion GPCR function.
Nat Commun, 15, 2024
|
|
6VI1
 
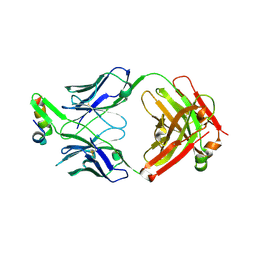 | | Structure of Fab4 bound to P22 TerL(1-33) | | Descriptor: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | Authors: | Cingolani, G, Lokareddy, R, Ko, Y. | | Deposit date: | 2020-01-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5CJO
 
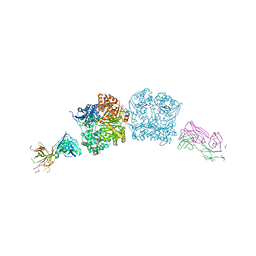 | |
7KEO
 
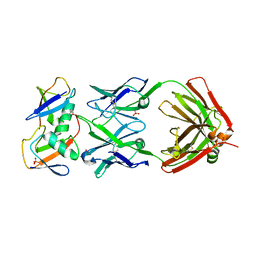 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7MDJ
 
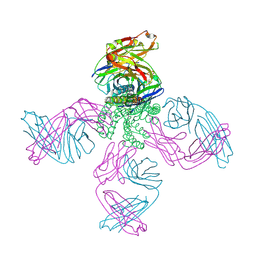 | | The structure of KcsA in complex with a synthetic Fab | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Slezak, T, Blackowicz, L, Kossiakoff, A, Roux, B. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering of a synthetic antibody fragment for structural and functional studies of K+ channels.
J.Gen.Physiol., 154, 2022
|
|
9PTI
 
 | |
7RU6
 
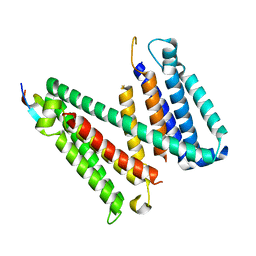 | |
7RUG
 
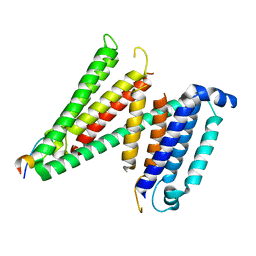 | | Human SERINC3-DeltaICL4 | | Descriptor: | Serine incorporator 3, SiA | | Authors: | Purdy, M.D, Leonhardt, S.A, Yeager, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry.
Nat Commun, 14, 2023
|
|
7TDN
 
 | |
7TDM
 
 | |
7T9X
 
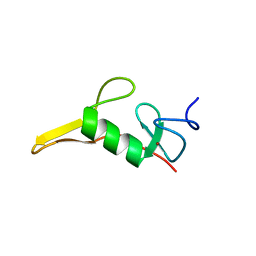 | | Saccharomyces cerevisiae Pex12 RING domain | | Descriptor: | Peroxisome assembly protein 12, ZINC ION | | Authors: | Feng, P, Rapoport, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel.
Nature, 607, 2022
|
|
