7FIM
 
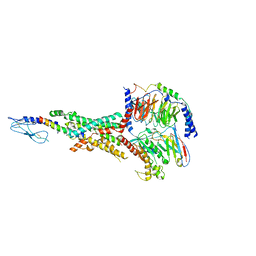 | | Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,human glucagon like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7T1F
 
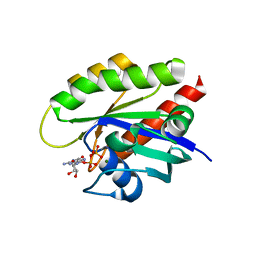 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
4DOY
 
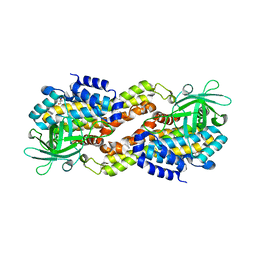 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
4GO3
 
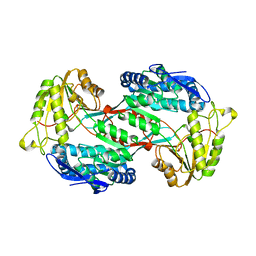 | |
4GO4
 
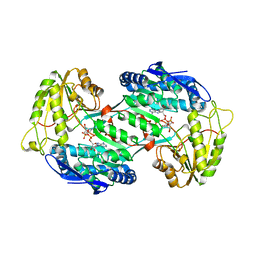 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
4HQX
 
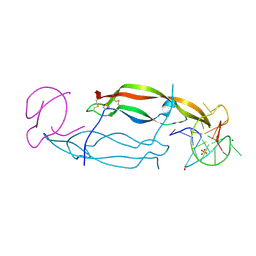 | | CRYSTAL STRUCTURE OF HUMAN PDGF-BB IN COMPLEX WITH A Modified nucleotide aptamer (SOMAmer SL4) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQU
 
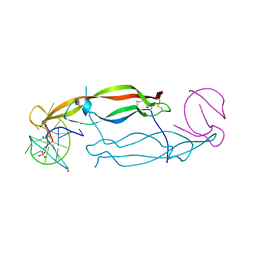 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3D9H
 
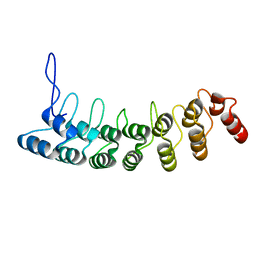 | | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein | | Descriptor: | cDNA FLJ77766, highly similar to Homo sapiens ankyrin repeat and SOCS box-containing 9 (ASB9), transcript variant 2, ... | | Authors: | Fei, X, Gu, X, Fan, S, Zhang, C, Ji, C. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein
To be published
|
|
3Q7J
 
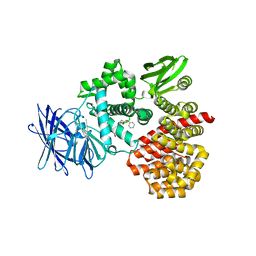 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
3R5T
 
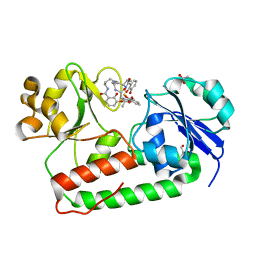 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3R5S
 
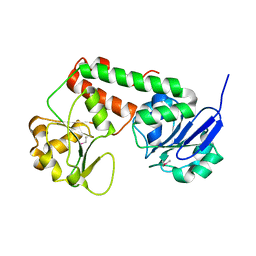 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
9B5Y
 
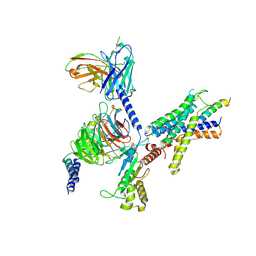 | | Cryo-EM structure of the LAPTH-bound PTH1R in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q), ... | | Authors: | Zhang, X, Liu, H, Zhang, C, Vilardaga, J.-P. | | Deposit date: | 2024-03-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric mechanism in the distinctive coupling of G q and G s to the parathyroid hormone type 1 receptor.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8W72
 
 | |
8W6Z
 
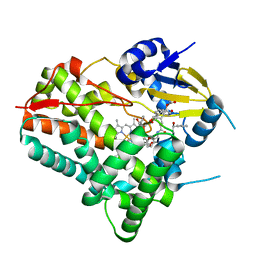 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Biosynthesis of Cihanmycins Reveal Cytochrome P450-Catalyzed Intramolecular C-O Phenol Coupling Reactions.
J.Am.Chem.Soc., 2024
|
|
8G59
 
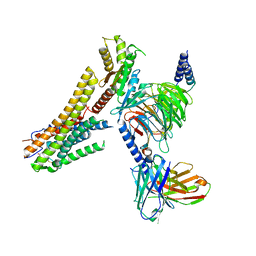 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
9JE0
 
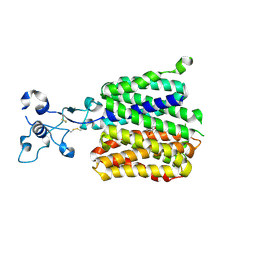 | | Human URAT1 bound to benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of urate transport by the native human URAT1 and its inhibition by anti-gout drugs.
Cell Discov, 11, 2025
|
|
9JDV
 
 | | Human URAT1 bound with Uric acid | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular mechanisms of urate transport by the native human URAT1 and its inhibition by anti-gout drugs.
Cell Discov, 11, 2025
|
|
9JDY
 
 | | Human URAT1 bound with verinurad | | Descriptor: | Solute carrier family 22 member 12, verinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular mechanisms of urate transport by the native human URAT1 and its inhibition by anti-gout drugs.
Cell Discov, 11, 2025
|
|
9JE1
 
 | | Human URAT1 bound to dotinurad | | Descriptor: | Solute carrier family 22 member 12, dotinurad | | Authors: | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of urate transport by the native human URAT1 and its inhibition by anti-gout drugs.
Cell Discov, 11, 2025
|
|
7LJC
 
 | | Allosteric modulator LY3154207 binding to SKF-81297-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7LJD
 
 | | Allosteric modulator LY3154207 binding to dopamine-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7NFA
 
 | | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer | | Descriptor: | A33 Fab heavy chain, A33 Fab light chain | | Authors: | Tang, J, Zhang, C, Dalby, P, Kozielski, F. | | Deposit date: | 2021-02-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer
To Be Published
|
|
5Z3Q
 
 | | Crystal Structure of a Soluble Fragment of Poliovirus 2C ATPase (2.55 Angstrom) | | Descriptor: | PHOSPHATE ION, PV-2C, ZINC ION | | Authors: | Guan, H, Tian, J, Zhang, C, Qin, B, Cui, S. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Crystal structure of a soluble fragment of poliovirus 2CATPase
PLoS Pathog., 14, 2018
|
|
