4IOP
 
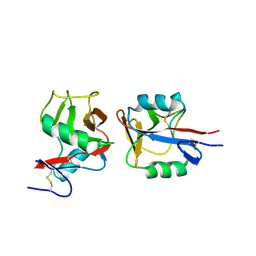 | | Crystal structure of NKp65 bound to its ligand KACL | | Descriptor: | C-type lectin domain family 2 member A, Killer cell lectin-like receptor subfamily F member 2, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y. | | Deposit date: | 2013-01-08 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Z2T
 
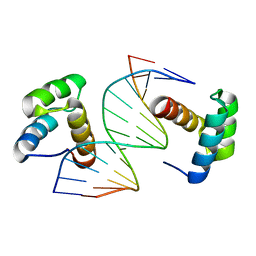 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
4KJU
 
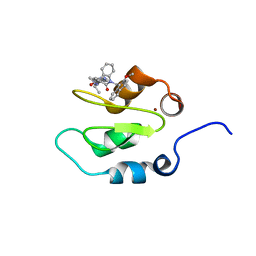 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
1X6M
 
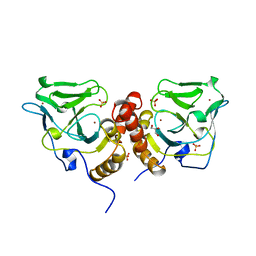 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
1VR0
 
 | |
1VQ3
 
 | |
1VR8
 
 | |
6DCW
 
 | |
5XLR
 
 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
7C2V
 
 | | Crystal Structure of IRAK4 kinase in complex with the inhibitor CA-4948 | | Descriptor: | 2-(2-methylpyridin-4-yl)-N-[2-morpholin-4-yl-5-[(3R)-3-oxidanylpyrrolidin-1-yl]-[1,3]oxazolo[4,5-b]pyridin-6-yl]-1,3-oxazole-4-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Krishnamurthy, N.R, Robert, B. | | Deposit date: | 2020-05-09 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of CA-4948, an Orally Bioavailable IRAK4 Inhibitor for Treatment of Hematologic Malignancies.
Acs Med.Chem.Lett., 11, 2020
|
|
7C2W
 
 | |
3HYB
 
 | |
6IE9
 
 | | RamR in complex with chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
6E4A
 
 | | Crystal structure of human BRD4(1) in complex with CN750 | | Descriptor: | 5-(4-{[(3-chlorophenyl)methyl]amino}-2-{4-[2-(dimethylamino)ethyl]piperazin-1-yl}quinazolin-6-yl)-1-methylpyridin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Fontano, E, White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and lead identification of quinazoline-based BRD4 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3HWN
 
 | | CATHEPSIN L with AZ13010160 | | Descriptor: | Cathepsin L1, Nalpha-[(3-tert-butyl-1-methyl-1H-pyrazol-5-yl)carbonyl]-N-[(2E)-2-iminoethyl]-3-{5-[(Z)-iminomethyl]-1,3,4-oxadiazol-2-yl}-L-phenylalaninamide | | Authors: | Kenny, P, Morley, A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-09-15 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Design of selective Cathepsin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6EEL
 
 | |
4QO2
 
 | | Crystal structure of rhomboid intramembrane protease GlpG in complex with peptide derived inhibitor Ac-IATA-cmk | | Descriptor: | 6-AMINO-2-METHYL-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, CHLORIDE ION, Rhomboid protease GlpG, ... | | Authors: | Zoll, S, Strisovsky, K. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures.
Embo J., 33, 2014
|
|
6EII
 
 | | The crystal structure of CK2alpha in complex with compound 18 | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-(3-phenylpropyl)azanium, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
6EHK
 
 | | The crystal structure of CK2alpha in complex with CAM4712 and compound 37 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[3,5-bis(chloranyl)-4-(2-ethylphenyl)phenyl]methyl]ethanamine, 2-hydroxy-5-methylbenzoic acid, ACETATE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6EHU
 
 | | The crystal structure of CK2alpha in complex with compound 32 | | Descriptor: | 2-(1~{H}-benzimidazol-2-yl)-~{N}-[[4-(2-ethylphenyl)-3-(trifluoromethyl)phenyl]methyl]ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-09-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6IE8
 
 | | RamR in complex with cholic acid | | Descriptor: | CHOLIC ACID, Regulatory protein, SULFATE ION | | Authors: | Nakashima, R, Sakurai, K, Yamasaki, S, Nishino, K. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the multidrug resistance regulator RamR complexed with bile acids.
Sci Rep, 9, 2019
|
|
4QO0
 
 | |
6F25
 
 | | Crystal structure of human acetylcholinesterase in complex with C35. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dias, J, Nachon, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05199647 Å) | | Cite: | New evidence for dual binding site inhibitors of acetylcholinesterase as improved drugs for treatment of Alzheimer's disease.
Neuropharmacology, 155, 2019
|
|
6FLG
 
 | |
