7V2S
 
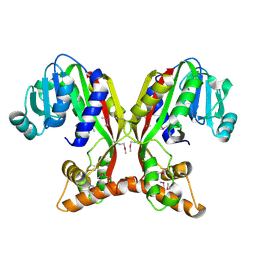 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT isoform3 from silkworm | | Descriptor: | Methyltranfer_dom domain-containing protein | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
9CS0
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP1 | | Descriptor: | Cyclic peptide CP1, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS1
 
 | | E. coli BamA beta-barrel bound to darobactin and cyclic peptide CP2 | | Descriptor: | Cyclic peptide CP2, Darobactin A, Outer membrane protein assembly factor BamA, ... | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
9CS2
 
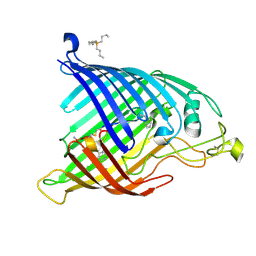 | | E. coli BamA beta-barrel bound to cyclic peptide CP3 | | Descriptor: | Cyclic peptide CP3, Outer membrane protein assembly factor BamA, tetrabutylphosphonium | | Authors: | Walker, M.E, Gu, M, Lu, J, Klein, D.J. | | Deposit date: | 2024-07-23 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Nat Commun, 16, 2025
|
|
8K0D
 
 | |
8K0C
 
 | |
3BJI
 
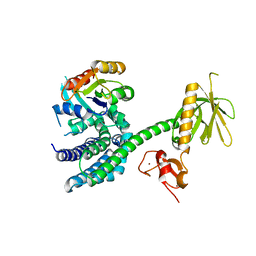 | | Structural Basis of Promiscuous Guanine Nucleotide Exchange by the T-Cell Essential Vav1 | | Descriptor: | Proto-oncogene vav, Ras-related C3 botulinum toxin substrate 1 precursor, ZINC ION | | Authors: | Chrencik, J.E, Brooun, A, Kuhn, P, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-04 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1.
J.Mol.Biol., 380, 2008
|
|
8IKL
 
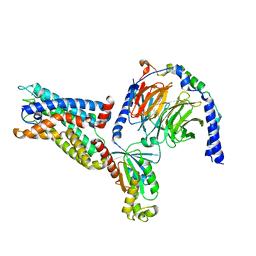 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8XJV
 
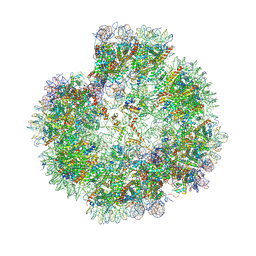 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
8UD9
 
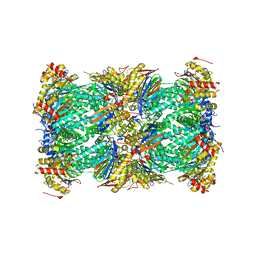 | | Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
7XR6
 
 | | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Zhao, Y, Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ligand binding modes of human EAAT2.
Nat Commun, 13, 2022
|
|
7XR4
 
 | |
1PHO
 
 | |
2DRI
 
 | |
5JQI
 
 | |
7XWO
 
 | | Neurokinin A bound to active human neurokinin 2 receptor in complex with G324 | | Descriptor: | G324, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W.J, Yuan, Q.N, Zhang, H.H, Yang, F, Ling, S.L, Lv, P, Eric, X, Tian, C.L, Yin, W.C, Shi, P. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the activation of neurokinin 2 receptor by neurokinin A.
Cell Discov, 8, 2022
|
|
6J9O
 
 | |
7W7F
 
 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7W77
 
 | | cryo-EM structure of human NaV1.3/beta1/beta2-bulleyaconitineA | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
8JA0
 
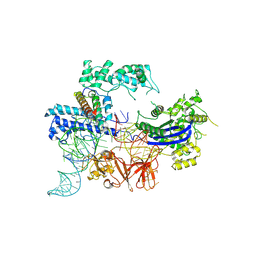 | | Cryo-EM structure of the NmeCas9-sgRNA-AcrIIC4 ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (117-MER), Uncharacterized protein | | Authors: | Yin, H, Li, Z, Yu, G.M, Li, X.Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8SUO
 
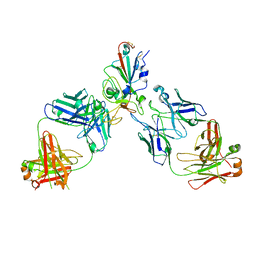 | | BA.2/AZD1061/AZD3152 structure analysis | | Descriptor: | AZD1061 heavy chain, AZD1061 light chain, AZD3152 heavy chain, ... | | Authors: | Oganesyan, V, van Dyk, N, Dippel, A, Barnes, A, O'Connor, E. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | AZD3152 neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults.
Sci Transl Med, 16, 2024
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPU
 
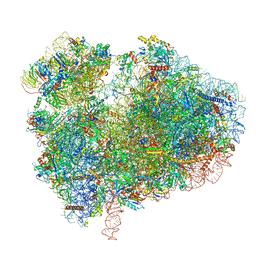 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7TX7
 
 | |
