7W3V
 
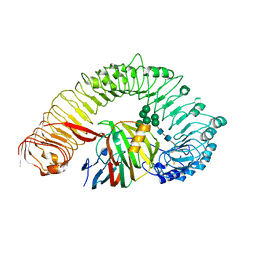 | | Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3T
 
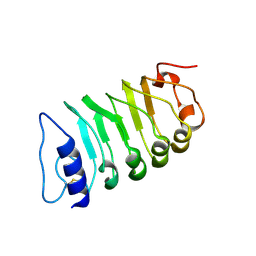 | | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
6CDY
 
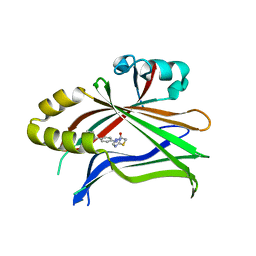 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
4PSX
 
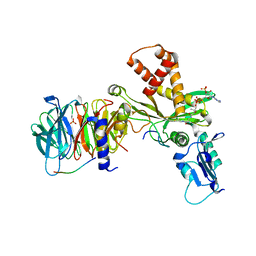 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY2
 
 | | Cryo-EM structure of apo MlaFEDB | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, Toluene tolerance protein Ttg2A, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY4
 
 | | Cryo-EM structure of MlaFEDB in complex with ADP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY9
 
 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4O1I
 
 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
7V6G
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
4O1H
 
 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2R7B
 
 | | Crystal Structure of the Phosphoinositide-dependent Kinase-1 (PDK-1)Catalytic Domain bound to a dibenzonaphthyridine inhibitor | | Descriptor: | 10,11-dimethoxy-4-methyldibenzo[c,f]-2,7-naphthyridine-3,6-diamine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2007-09-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Dibenzo[c,f][2,7]naphthyridines as Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 Inhibitors.
J.Med.Chem., 50, 2007
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
3LPC
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | ACETATE ION, AprB2, CALCIUM ION, ... | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LPA
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LPD
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
5DBF
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NADPH | | Descriptor: | Iridoid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1P5T
 
 | | Crystal Structure of Dok1 PTB Domain | | Descriptor: | Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yuan, J, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.BIOL.CHEM., 279, 2004
|
|
6KUJ
 
 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 1 | | Descriptor: | 3'-cRNA promoter, 5'-cRNA promoter, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of influenza D virus polymerase bound to cRNA promoter in Mode A conformation
NAT NANOTECHNOL, 2019
|
|
5DBI
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ and 10-oxogeranial | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
