4S1Q
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1S
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H5.F-185917, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab of VRC01 light chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
5YGE
 
 | | ArgA complexed with AceCoA and glutamate | | Descriptor: | ACETYL COENZYME *A, Amino-acid acetyltransferase, CACODYLIC ACID, ... | | Authors: | Yang, X, Wu, L, Ran, Y, Xu, A, Zhang, B, Yang, X, Zhang, R, Rao, Z, Li, J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Crystal structure of l-glutamate N-acetyltransferase ArgA from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1865, 2017
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
3I9N
 
 | | Crystal structure of human CD38 complexed with an analog ribo-2'F-ADP ribose | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9K
 
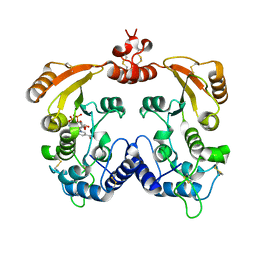 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9M
 
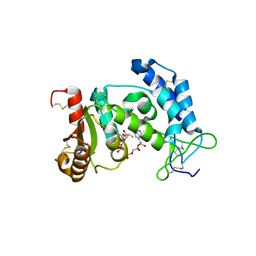 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9L
 
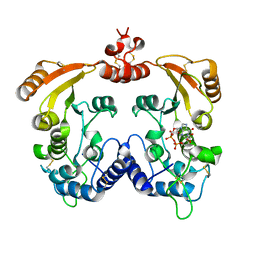 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
3I9J
 
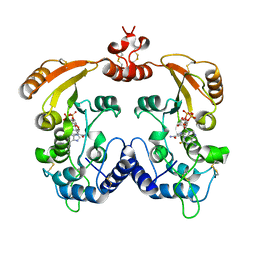 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
4GKY
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4201 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
4GKX
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4R7A
 
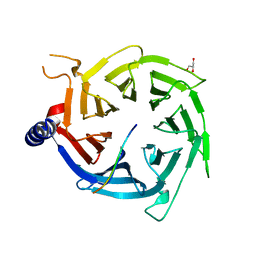 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | Descriptor: | Llama VHH A12, SULFATE ION | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-03-08 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
8HXQ
 
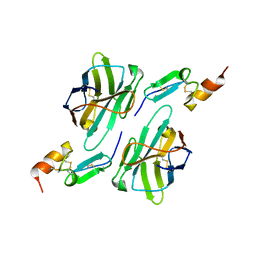 | | Nanobody1 in complex with human BCMA ECD | | Descriptor: | Nanobody1, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
8HXR
 
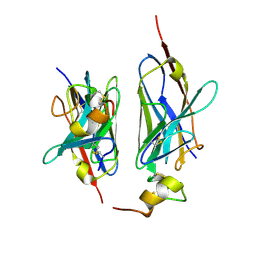 | | Nanobody2 in complex with human BCMA ECD | | Descriptor: | Nanobody2, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
5YC4
 
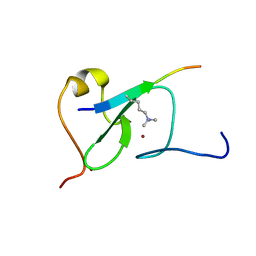 | |
5YC3
 
 | |
8GTY
 
 | |
6XVH
 
 | | carbonic anhydrase with bound arylsulfonamide- boronic acid | | Descriptor: | (4-sulfamoylphenyl)boronic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Perez, C, Zhang, B. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An easy to assemble, fully modulable, selectiv and
colorimetric artificial bio-sensor for fingerprintting
the recognition of neurotransmitters
To Be Published
|
|
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
5K6C
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.576 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5K6H
 
 | |
5K6I
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 A149C-Y458C, S46G-E92D-S215P-K465Q variant. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
