1BI8
 
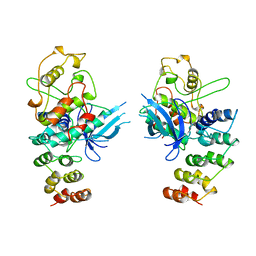 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1CA4
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 (TRAF2) | | Descriptor: | PROTEIN (TNF RECEPTOR ASSOCIATED FACTOR 2) | | Authors: | Park, Y.C, Burkitt, V, Villa, A.R, Tong, L, Wu, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for self-association and receptor recognition of human TRAF2.
Nature, 398, 1999
|
|
6WUF
 
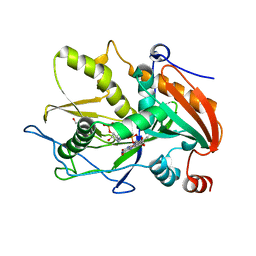 | | Crystal structure of mouse DXO in complex with 3'-FADP | | Descriptor: | 1,2-ETHANEDIOL, Decapping and exoribonuclease protein, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
6WRE
 
 | |
6X7V
 
 | |
6X7U
 
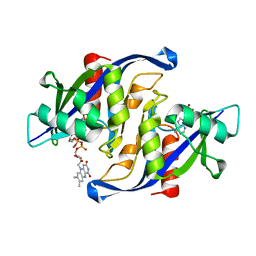 | |
6XGT
 
 | |
6XNV
 
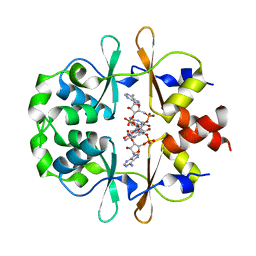 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES CBPB PROTEIN (LMO1009) IN COMPLEX WITH C-DI-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (p)ppGpp and c-di-AMP Homeostasis Is Controlled by CbpB in Listeria monocytogenes.
Mbio, 11, 2020
|
|
6XNU
 
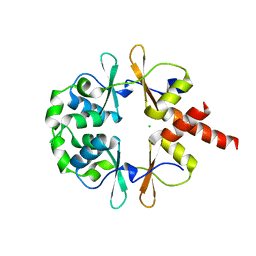 | |
6WUK
 
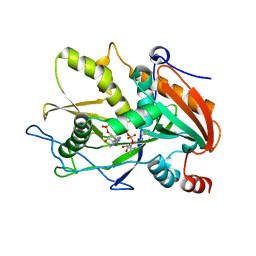 | |
6WUG
 
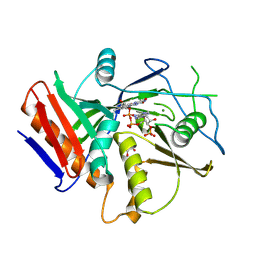 | |
6WUI
 
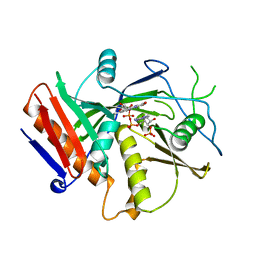 | | Crystal Structure of mutant S. pombe Rai1 (E150S/E199Q/E239Q) in complex with 3'-FADP | | Descriptor: | Decapping nuclease din1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
1JQ7
 
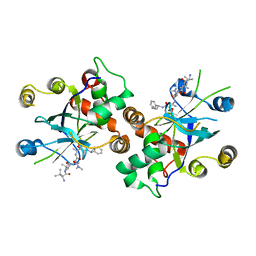 | | HCMV protease dimer-interface mutant, S225Y complexed to Inhibitor BILC 408 | | Descriptor: | ASSEMBLIN, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Batra, R, Khayat, R, Tong, L. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease.
Nat.Struct.Biol., 8, 2001
|
|
1IEG
 
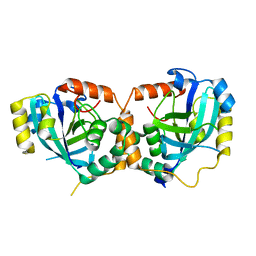 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A/H157A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IEF
 
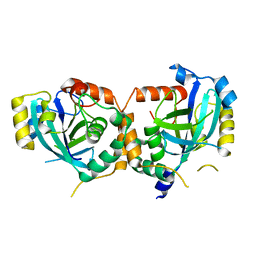 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT S134A OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
1IED
 
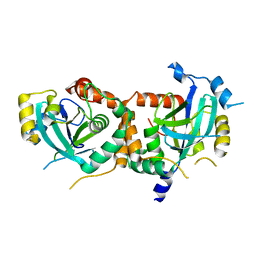 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157E) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
6URG
 
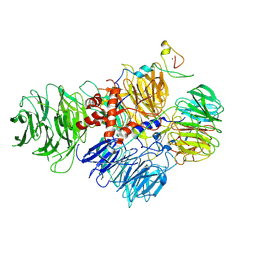 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
8E3Q
 
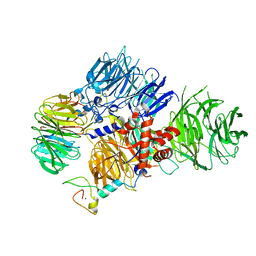 | | CRYO-EM STRUCTURE OF the human MPSF | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, ZINC ION, ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
6URO
 
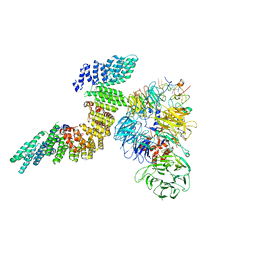 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6V4X
 
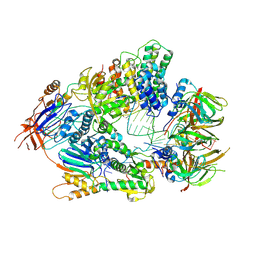 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
8EJM
 
 | |
1ID4
 
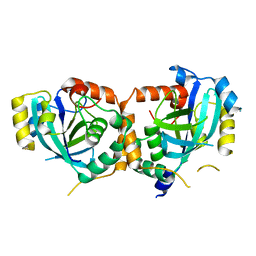 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157Q) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | Authors: | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | Deposit date: | 2001-04-03 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
8E3I
 
 | | CRYO-EM STRUCTURE OF the human MPSF IN COMPLEX WITH THE AUUAAA poly(A) signal | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*CP*AP*UP*UP*AP*AP*AP*CP*AP*AP*C)-3'), ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
6UN9
 
 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
5BTE
 
 | |
