8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRD
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRH
 
 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
 | |
8H68
 
 | | Crystal structure of Caenorhabditis elegans NMAD-1 in complex with NOG and Mg(II) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shi, Y, Ding, J, Yang, H. | | Deposit date: | 2022-10-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caenorhabditis elegans NMAD-1 functions as a demethylase for actin.
J Mol Cell Biol, 15, 2023
|
|
3NFS
 
 | | Crystal structure the Fab fragment of therapeutic antibody daclizumab | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
3NFP
 
 | | Crystal structure of the Fab fragment of therapeutic antibody daclizumab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of daclizumab, Interleukin-2 receptor subunit alpha, Light chain of Fab fragment of daclizumab | | Authors: | Yang, H, Wang, J, Du, J, Zhong, C, Guo, Y, Ding, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis of immunosuppression by the therapeutic antibody daclizumab
Cell Res., 20, 2010
|
|
2P6E
 
 | |
2OSL
 
 | | Crystal structure of Rituximab Fab in complex with an epitope peptide | | Descriptor: | B-lymphocyte antigen CD20, heavy chain of the Rituximab Fab fragment,heavy chain of the Rituximab Fab fragment, light chain of the Rituximab Fab fragment,light chain of the Rituximab Fab fragment | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of CD20 by therapeutic antibody Rituximab
J.Biol.Chem., 282, 2007
|
|
2P6G
 
 | |
2P6F
 
 | |
3CXD
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with its epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Yang, H, Zhong, C, Ding, J. | | Deposit date: | 2008-04-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | Descriptor: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | Authors: | Du, J, Zhong, C, Yang, H, Ding, J. | | Deposit date: | 2008-07-12 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3OPT
 
 | | Crystal structure of the Rph1 catalytic core with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DNA damage-responsive transcriptional repressor RPH1, NICKEL (II) ION | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OPW
 
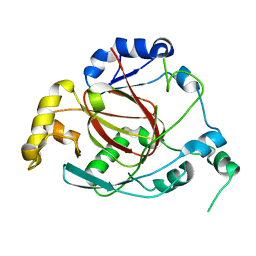 | | Crystal Structure of the Rph1 catalytic core | | Descriptor: | DNA damage-responsive transcriptional repressor RPH1 | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
5B7J
 
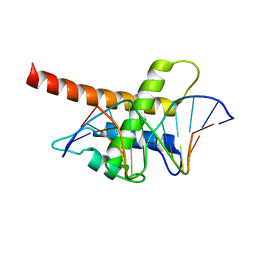 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
5EUE
 
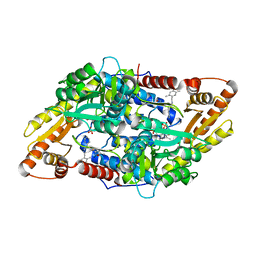 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
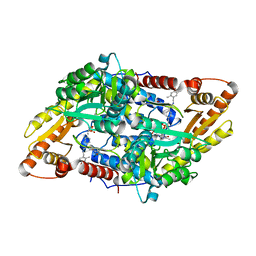 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1QK6
 
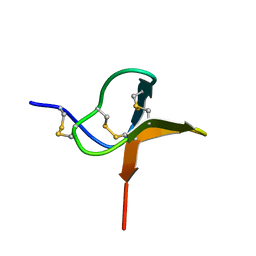 | | Solution structure of huwentoxin-I by NMR | | Descriptor: | HUWENTOXIN-I | | Authors: | Qu, Y, Liang, S, Ding, J, Liu, X, Zhang, R, Gu, X. | | Deposit date: | 1999-07-10 | | Release date: | 1999-08-20 | | Last modified: | 2019-01-16 | | Method: | SOLUTION NMR | | Cite: | Proton Nuclear Magnetic Resonance Studies on Huwentoxin-I from the Venom of the Spider Selenocosmia Huwena:2.Three-Dimensional Structure in Solution
J.Protein Chem., 16, 1997
|
|
5GK9
 
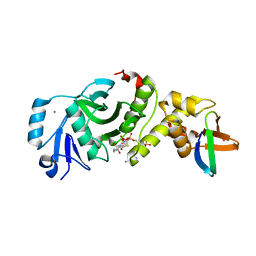 | | Crystal structure of human HBO1 in complex with BRPF2 | | Descriptor: | ACETYL COENZYME *A, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Tao, Y, Zhu, J, Xu, S, Ding, J. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2.
Nucleic Acids Res., 45, 2017
|
|
5H48
 
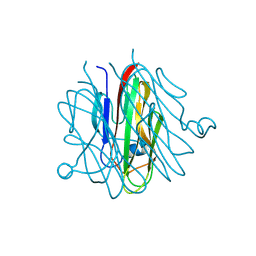 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H49
 
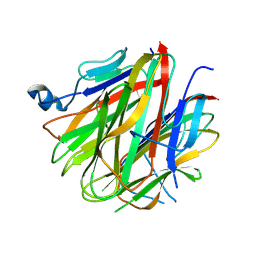 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
1O5T
 
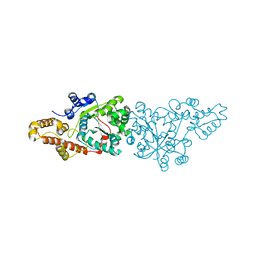 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
3OXL
 
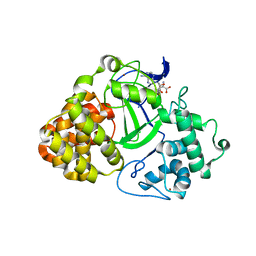 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form II) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding
Nucleic Acids Res., 39, 2011
|
|
