7MJR
 
 | | Vip4Da2 toxin structure | | Descriptor: | CALCIUM ION, SULFATE ION, Vip4Da1 protein | | Authors: | Rydel, T.J, Duda, D, Zheng, M, Henry, A. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional insights into the first Bacillus thuringiensis vegetative insecticidal protein of the Vpb4 fold, active against western corn rootworm.
Plos One, 16, 2021
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XJZ
 
 | | Structure of DNA1-Ag complex | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*GP*CP*GP*C)-3'), SILVER ION, SPERMINE | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6A85
 
 | | Crystal structure of a novel DNA quadruplex | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*GP*GP*GP*TP*GP*CP*GP*TP*T)-3'), LEAD (II) ION, ... | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2018-07-06 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads.
Nucleic Acids Res., 46, 2018
|
|
2K8H
 
 | |
4DNW
 
 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
7RCO
 
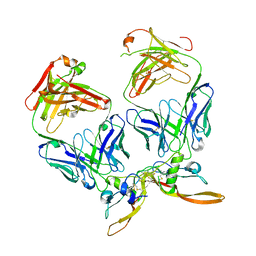 | |
2JZ3
 
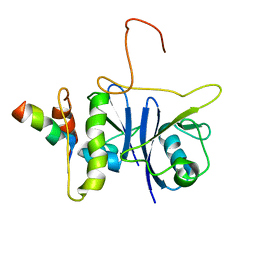 | | SOCS box elonginBC ternary complex | | Descriptor: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | Deposit date: | 2007-12-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
3QDQ
 
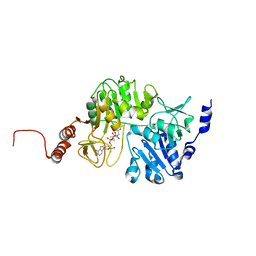 | |
6CFD
 
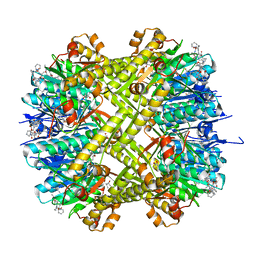 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7WN1
 
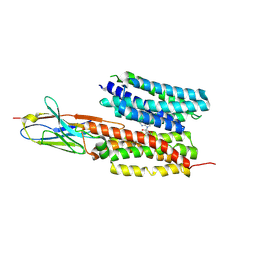 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
3ROC
 
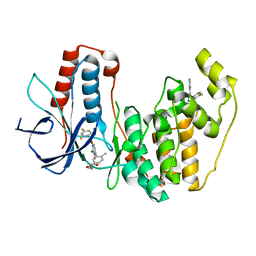 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4DUS
 
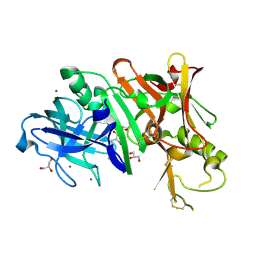 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
4OB6
 
 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | Descriptor: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
6VCH
 
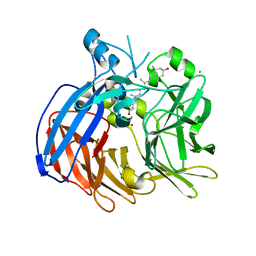 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase in complex with 3-hydroxy-beta-apo-14'-carotenal | | Descriptor: | (2E,4E,6E,8E,10E)-11-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-5,9-dimethylundeca-2,4,6,8,10-pentaenal, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4OB7
 
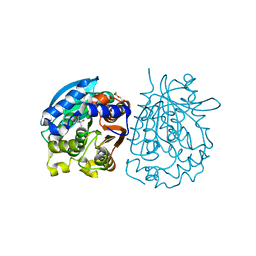 | | Crystal structure of esterase rPPE mutant W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-01-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
4OU5
 
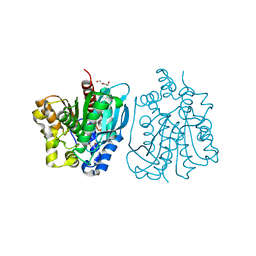 | | Crystal structure of esterase rPPE mutant S159A/W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
6VCF
 
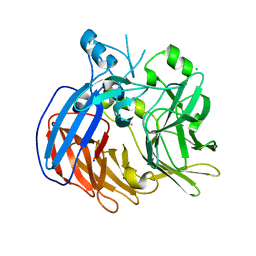 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, iron form | | Descriptor: | BICARBONATE ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.687 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VHS
 
 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | Descriptor: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M.D, Chen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
7MWH
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the BAZ2B TAM domain.
Heliyon, 8, 2022
|
|
5ZHE
 
 | | STRUCTURE OF E. COLI UNDECAPRENYL DIPHOSPHATE SYNTHASE IN COMPLEX WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
