4JAI
 
 | | Crystal Structure of Aurora Kinase A in complex with N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Descriptor: | Aurora kinase A, N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
4L4M
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4JAJ
 
 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5HP7
 
 | | Crystal structures of RidA in the apo form | | Descriptor: | Reactive Intermediate Deaminase A, chloroplastic | | Authors: | Xie, W, Liu, X. | | Deposit date: | 2016-01-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of RidA, an important enzyme for the prevention of toxic side products
Sci Rep, 6, 2016
|
|
4L4L
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.122 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2018-12-19 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
7D32
 
 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D31
 
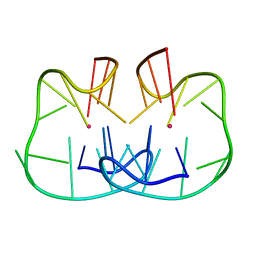 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D33
 
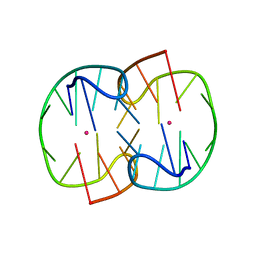 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
8SPM
 
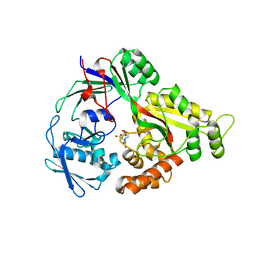 | | Crystal structure of NikA in complex Ni-AMA | | Descriptor: | Aspergillomarasmine A, NICKEL (II) ION, Nickel ABC transporter, ... | | Authors: | Sychantha, D, Prehna, G, Wright, G.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting bacterial nickel transport with aspergillomarasmine A suppresses virulence-associated Ni-dependent enzymes.
Nat Commun, 15, 2024
|
|
8T3P
 
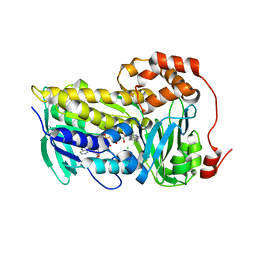 | |
8TCI
 
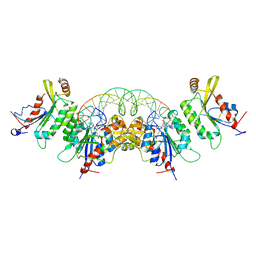 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | Descriptor: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | Authors: | Khudaverdyan, N, Song, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 2024
|
|
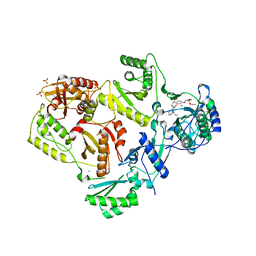
4I7F
 
 | |
2I4W
 
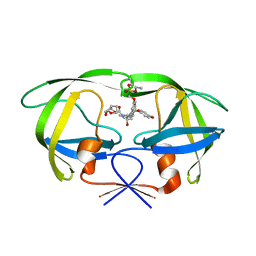 | | HIV-1 protease WT with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4U
 
 | | HIV-1 protease with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4D
 
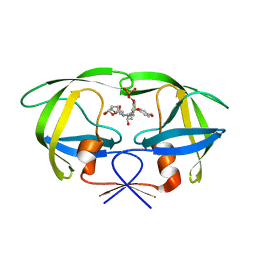 | | Crystal structure of WT HIV-1 protease with GS-8373 | | Descriptor: | ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONIC ACID, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-21 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4V
 
 | | HIV-1 protease I84V, L90M with TMC126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
5X0T
 
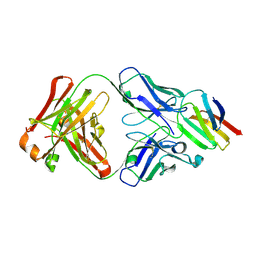 | | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab fragment heavy chain, 6H8 Fab fragment light chain, Basigin | | Authors: | Lin, P, Zhang, M.-Y, Ye, S, Chen, X, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
2I4X
 
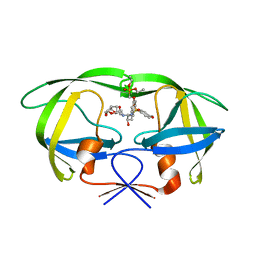 | | HIV-1 Protease I84V, L90M with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
6M6I
 
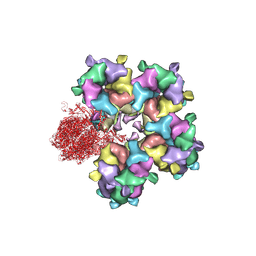 | | Structure of HSV2 B-capsid portal vertex | | Descriptor: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
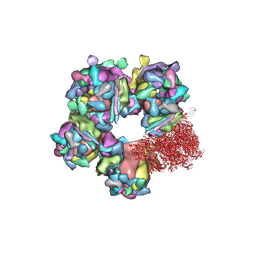 | | Structure of HSV2 C-capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
 | | Structure of HSV2 viron capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.39 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6VLB
 
 | |
