7CFS
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CFT
 
 | | Cryo-EM strucutre of human acid-sensing ion channel 1a in complex with snake toxin Mambalgin1 at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Mambalgin-1 | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
6O1S
 
 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
7CN8
 
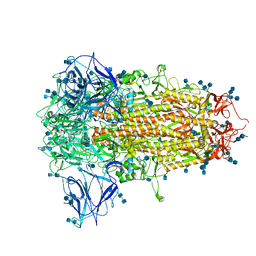 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
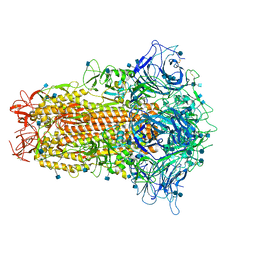 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
6ONU
 
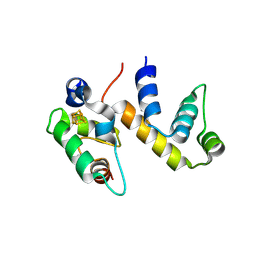 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
5BPW
 
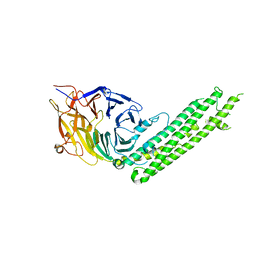 | |
5BPZ
 
 | | Atomic-resolution structures of the APC/C subunits Apc4 and the Apc5 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Anapc5 protein | | Authors: | Cronin, N, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Atomic-Resolution Structures of the APC/C Subunits Apc4 and the Apc5 N-Terminal Domain.
J.Mol.Biol., 427, 2015
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
5LGG
 
 | | The N-terminal WD40 domain of Apc1 (Anaphase promoting complex subunit 1) | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1 | | Authors: | Li, Q, Aibara, S, Barford, D. | | Deposit date: | 2016-07-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WD40 domain of Apc1 is critical for the coactivator-induced allosteric transition that stimulates APC/C catalytic activity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8GZ8
 
 | | Cryo-EM structure of Abeta2 fibril polymorph1 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8GZ9
 
 | | Cryo-EM structure of Abeta2 fibril polymorph2 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
4R91
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(cyclopentylamino)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(1S,3R)-3-(cyclopentylamino)cyclohexyl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R95
 
 | | BACE-1 in complex with 2-(((1R,3S)-3-(((R)-4-(2-cyclohexylethyl)-2-iminio-1-methyl-5-oxoimidazolidin-4-yl)methyl)cyclohexyl)amino)quinolin-1-ium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-2-imino-3-methyl-5-{[(1S,3R)-3-(quinolin-2-ylamino)cyclohexyl]methyl}imidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R92
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R8Y
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((R)-1-(2-cyclopentylacetyl)pyrrolidin-3-yl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-{[(3R)-1-(cyclopentylacetyl)pyrrolidin-3-yl]methyl}-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Caldwell, J.P, Strickland, C. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4R93
 
 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-1-methyl-5-oxo-4-(((1S,3R)-3-(3-phenylureido)cyclohexyl)methyl)imidazolidin-2-iminium | | Descriptor: | 1-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]-3-phenylurea, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
