5XQ1
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7F0H
 
 | |
4QN7
 
 | | Crystal structure of neuramnidase N7 complexed with Oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4I7F
 
 | |
6IZO
 
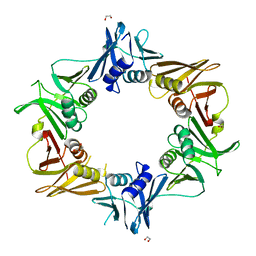 | | Crystal structure of DNA polymerase sliding clamp from Caulobacter crescentus | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER | | Authors: | Jiang, X, Zhang, L, Teng, M, Li, X. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
4QN5
 
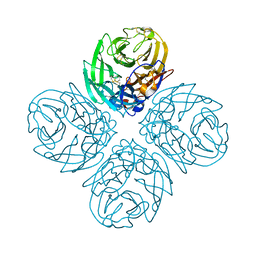 | | Neuraminidase N5 binds LSTa at the second SIA binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4QN6
 
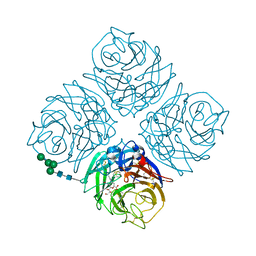 | | Crystal structure of Neuraminidase N6 complexed with Laninamivir | | Descriptor: | 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, Neuraminidase, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
6JIR
 
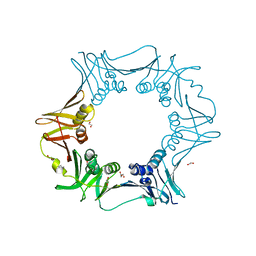 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
1A4V
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chandra, N, Acharya, K.R. | | Deposit date: | 1998-02-05 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for the presence of a secondary calcium binding site in human alpha-lactalbumin.
Biochemistry, 37, 1998
|
|
4QN4
 
 | | Crystal structure of Neuraminidase N6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4KPS
 
 | | Structure and receptor binding specificity of the hemagglutinin H13 from avian influenza A virus H13N6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Lu, X, Qi, J, Shi, Y, Gao, G. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structure and Receptor Binding Specificity of Hemagglutinin H13 from Avian Influenza A Virus H13N6
J.Virol., 87, 2013
|
|
4KPQ
 
 | | Structure and receptor binding specificity of the hemagglutinin H13 from avian influenza A virus H13N6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Qi, J, Shi, Y, Gao, G. | | Deposit date: | 2013-05-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and Receptor Binding Specificity of Hemagglutinin H13 from Avian Influenza A Virus H13N6
J.Virol., 87, 2013
|
|
4QN3
 
 | | Crystal structure of Neuraminidase N7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | Descriptor: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
8WIE
 
 | | Peptide 10-1/FTH1-1 Complex | | Descriptor: | CALCIUM ION, FE (III) ION, Peptide 10-1,Ferritin heavy chain | | Authors: | Fu, D, Wang, M, Guo, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-09-25 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Self-assembling nanoparticle engineered from the ferritinophagy complex as a rabies virus vaccine candidate.
Nat Commun, 15, 2024
|
|
8WJF
 
 | | Peptide 10/FTH1 complex | | Descriptor: | Peptide 10,Ferritin heavy chain | | Authors: | Fu, D, Wang, M, Guo, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Self-assembling nanoparticle engineered from the ferritinophagy complex as a rabies virus vaccine candidate.
Nat Commun, 15, 2024
|
|
8WIQ
 
 | | NCOA4/FTH1 complex | | Descriptor: | Native peptide,Ferritin heavy chain | | Authors: | Fu, D, Wang, M, Guo, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Self-assembling nanoparticle engineered from the ferritinophagy complex as a rabies virus vaccine candidate.
Nat Commun, 15, 2024
|
|
7XBW
 
 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with Gi1 | | Descriptor: | CHOLESTEROL, CX3C chemokine receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
7XBX
 
 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with CX3CL1 and Gi1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
8GS9
 
 | | SARS-CoV-2 BA.2 spike RBD in complex bound with VacBB-551 | | Descriptor: | Heavy chain of VacBB-551, Light chain of VacBB-551, Spike glycoprotein | | Authors: | Liu, C.C, Ju, B, Shen, S.L, Zhang, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron BQ.1.1 and XBB.1 unprecedentedly escape broadly neutralizing antibodies elicited by prototype vaccination.
Cell Rep, 42, 2023
|
|
8YPU
 
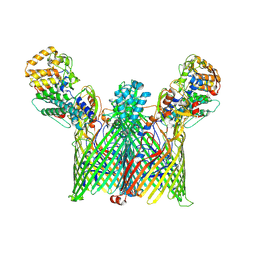 | | Cryo-EM structure of ButCD complex | | Descriptor: | Membrane protein, SusC/RagA family TonB-linked outer membrane protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
8YPT
 
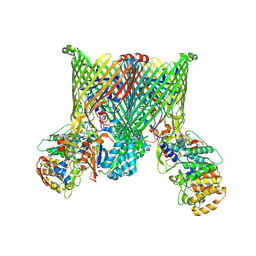 | | Cryo-EM structure of BfUbb-ButCD complex | | Descriptor: | Membrane protein, TonB-linked outer membrane protein, Ubiquitin-like protein | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2024-03-18 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A highly conserved SusCD transporter determines the import and species-specific antagonism of Bacteroides ubiquitin homologues.
Nat Commun, 15, 2024
|
|
7VRN
 
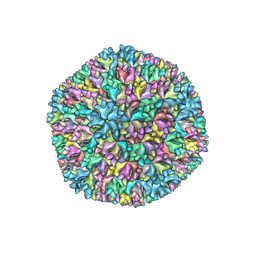 | | Structure of infectious bursal disease virus Gt strain | | Descriptor: | Structural polyprotein | | Authors: | Bao, K.Y, Zhu, P. | | Deposit date: | 2021-10-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of infectious bursal disease viruses with different virulences provide insights into their assembly and invasion
Sci Bull (Beijing), 67, 2022
|
|
7VRP
 
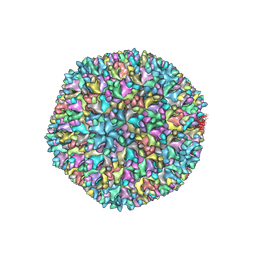 | | Structure of infectious bursal disease virus Gx strain | | Descriptor: | Structural polyprotein | | Authors: | Bao, K.Y, Zhu, P. | | Deposit date: | 2021-10-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of infectious bursal disease viruses with different virulences provide insights into their assembly and invasion
Sci Bull (Beijing), 67, 2022
|
|
7BEG
 
 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
