6DE3
 
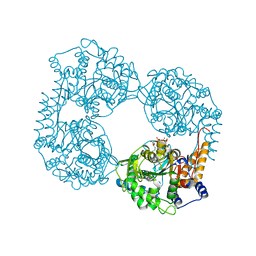 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-11 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
1Y2B
 
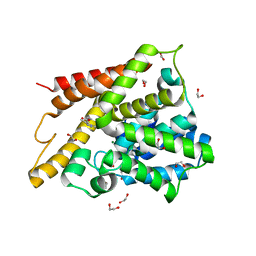 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
6DNZ
 
 | | Trypanosoma brucei PRMT1 enzyme-prozyme heterotetrameric complex with AdoHcy | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine N-methyltransferase, putative, ... | | Authors: | Hashimoto, H, Kafkova, L, Jordan, K, Read, L.K, Debler, E.W. | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Structural Basis of Protein Arginine Methyltransferase Activation by a Catalytically Dead Homolog (Prozyme).
J.Mol.Biol., 432, 2020
|
|
1YD3
 
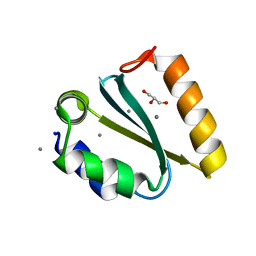 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y43F bound to its catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
5EHO
 
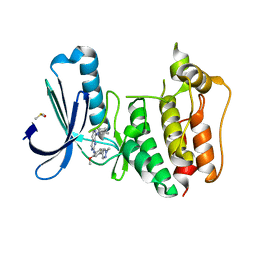 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}8-cyclohexyl-~{N}2-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
6D7Y
 
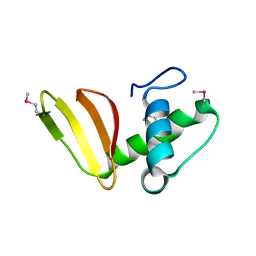 | | 1.75 Angstrom Resolution Crystal Structure of the Toxic C-Terminal Tip of CdiA from Pseudomonas aeruginosa in Complex with Immune Protein | | Descriptor: | Hemagglutinin, immune protein | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Allen, J.P, Hauser, A.R, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A comparative genomics approach identifies contact-dependent growth inhibition as a virulence determinant.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8OLL
 
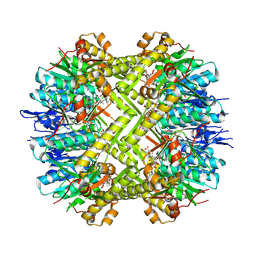 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLR
 
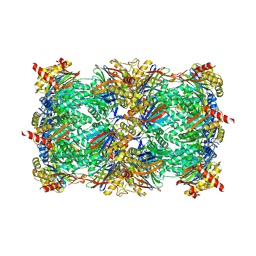 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6D0J
 
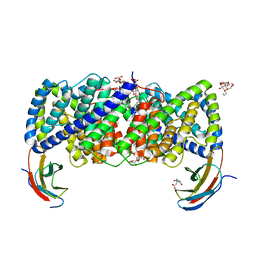 | | Crystal structure of a CLC-type fluoride/proton antiporter | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5EKP
 
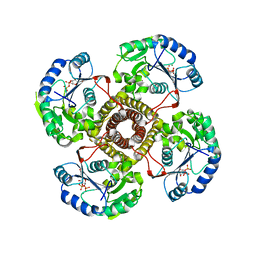 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
8P53
 
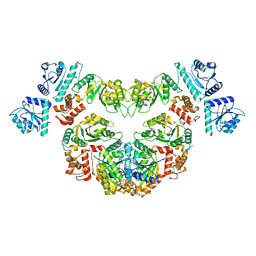 | |
5EMJ
 
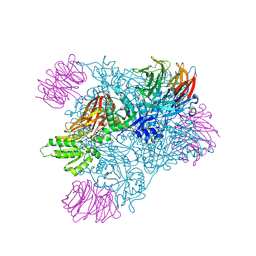 | | Crystal structure of PRMT5:MEP50 with Compound 8 and sinefungin | | Descriptor: | (2~{S})-1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-3-[[4-(3-methylbenzimidazol-5-yl)pyridin-2-yl]amino]propan-2-ol, GLYCEROL, Methylosome protein 50, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666.
ACS Med Chem Lett, 7, 2016
|
|
1Y6G
 
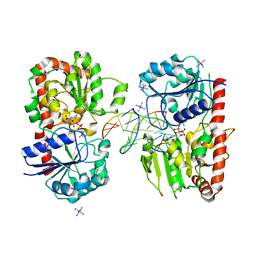 | | alpha-glucosyltransferase in complex with UDP and a 13_mer DNA containing a HMU base at 2.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*T)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(5HU)P*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
8OMR
 
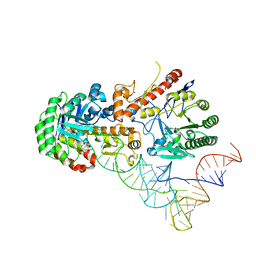 | | Human tRNA guanine transglycosylase (TGT) bound to tRNAAsp | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sievers, K, Neumann, P, Susac, L, Trowitzsch, S, Tampe, R, Ficner, R. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into tRNA recognition by human tRNA guanine transglycosylase.
Structure, 32, 2024
|
|
1YA6
 
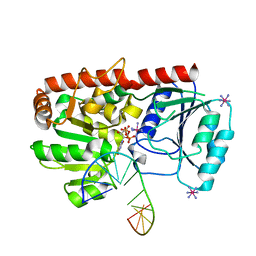 | | alpha-glucosyltransferase in complex with UDP and a 13-mer DNA containing a central A:G mismatch | | Descriptor: | 5'-D(*AP*TP*AP*CP*TP*AP*AP*GP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*T)-3', COBALT HEXAMMINE(III), ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
1URC
 
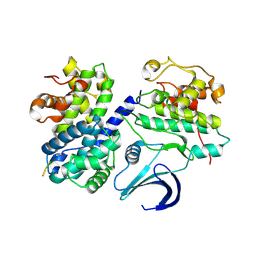 | | Cyclin A binding groove inhibitor Ace-Arg-Lys-Leu-Phe-Gly | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, PEPTIDE INHIBITOR | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-10-28 | | Release date: | 2003-10-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, biological activity and structural analysis of cyclic peptide inhibitors targeting the substrate recruitment site of cyclin-dependent kinase complexes.
Org. Biomol. Chem., 2, 2004
|
|
6CYA
 
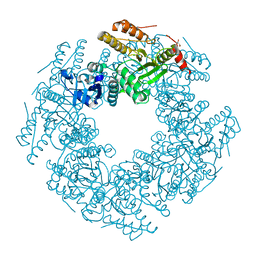 | | Rotavirus SA11 NSP2 S313A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 2 | | Authors: | Anish, R, Hu, L, Prasad, B.V.V. | | Deposit date: | 2018-04-05 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8OHQ
 
 | | Crystal structure of human heparanase in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J, Overkleeft, H.S, Armstrong, Z, Yurong, C. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHV
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
5EML
 
 | |
6CYK
 
 | | CTX-M-14 N106S mutant | | Descriptor: | ACETATE ION, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
8OHX
 
 | | Crystal structure of Beta-glucuronidase from Escherichia coli in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OGX
 
 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHT
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHU
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured isofagamine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
