8SRX
 
 | | Crystal structure of BAK-BAX heterodimer with lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, Bcl-2 homologous antagonist/killer, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-07 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SRY
 
 | | Crystal structure of BAK-BAX heterodimer with C12E8 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, ... | | Authors: | Brouwer, J.M, Czabotar, P.E, Colman, P.M, Miller, M.S. | | Deposit date: | 2023-05-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SDF
 
 | |
8SIT
 
 | |
8SDG
 
 | |
8SIS
 
 | |
8SIR
 
 | |
8SIQ
 
 | |
8SDH
 
 | |
8V08
 
 | |
8V06
 
 | | Crystal structure of mouse PLD3 co-crystallized with 5'Pi-ssDNA for 9 days | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
8V05
 
 | | Crystal structure of mouse PLD3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
8V07
 
 | | Crystal structure of mouse PLD3 co-crystallized with 5'Pi-ssDNA for 30 days | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
7NAB
 
 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
6ATT
 
 | | 39S Fab bound to HER2 ecd | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 39S Fab heavy chain, ... | | Authors: | Oganesyan, V.Y, Dall'Acqua, W.F. | | Deposit date: | 2017-08-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Structural insights into the mechanism of action of a biparatopic anti-HER2 antibody.
J. Biol. Chem., 293, 2018
|
|
7CCE
 
 | |
7ER2
 
 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with LS_2_40 | | Descriptor: | 5-chloranyl-N2-[3-chloranyl-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-N4-(2-dimethylphosphorylphenyl)pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2021-05-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Optimization of Brigatinib as New Wild-Type Sparing Inhibitors of EGFR T790M/C797S Mutants.
Acs Med.Chem.Lett., 13, 2022
|
|
7YFK
 
 | | The structure of human pregnane X receptor in complex with an SRC-1 coactivator peptide and a limonoid compound, nomilin | | Descriptor: | Nomilin, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 | | Authors: | Xia, Y, Yao, D, Huang, C, Cao, Y. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pregnane X receptor agonist nomilin extends lifespan and healthspan in preclinical models through detoxification functions.
Nat Commun, 14, 2023
|
|
8G1T
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8H3U
 
 | | Inhibitor-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
8H3S
 
 | | Substrate-bound EP, polyA model | | Descriptor: | Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain, Serine protease 1 | | Authors: | Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-10-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7XPJ
 
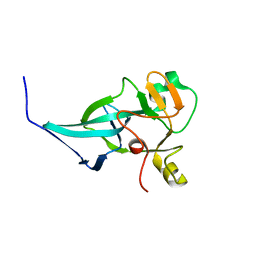 | | crystal structure of rice ASI1 BAH domain | | Descriptor: | BAH domain-containing protein | | Authors: | Yuan, J, Du, J. | | Deposit date: | 2022-05-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Molecular basis of locus-specific H3K9 methylation catalyzed by SUVH6 in plants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XPK
 
 | |
7YGG
 
 | |
7WQZ
 
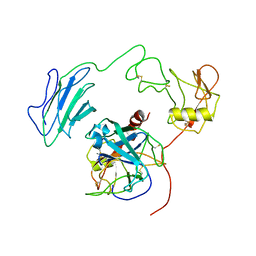 | | Structure of Active-mutEP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
