7KSP
 
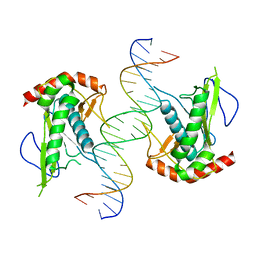 | | Crystal structure of hSAMD9_DBD with DNA | | Descriptor: | DNA, Sterile alpha motif domain-containing protein 9 | | Authors: | Peng, S, Pathak, P, Xiang, Y, Deng, J. | | Deposit date: | 2020-11-23 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of an effector domain in antiviral factors and tumor suppressors SAMD9 and SAMD9L.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ED3
 
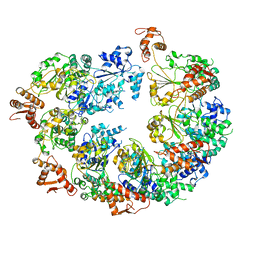 | | Mtb ClpB in complex with AMPPNP | | Descriptor: | Chaperone protein ClpB | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7XHO
 
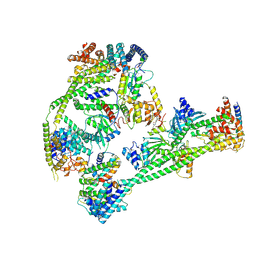 | | Structure of human inner kinetochore CCAN complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Tian, T, Wang, C.L, Yang, Z.S, Sun, L.F, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7XHN
 
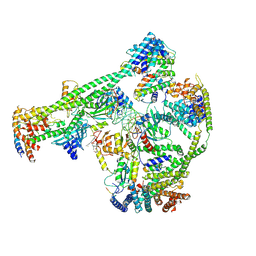 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
3KEA
 
 | |
4LU5
 
 | |
4LQF
 
 | | Structure of murine IgG2b A2C7-Fab in complex with vaccinia antigen A33R at the resolution of 2.3 Angstroms | | Descriptor: | A33R, Murine IgG2b A2C7 Heavy chain Fab domain, Murine IgG2b A2C7 Light chain Fab domain, ... | | Authors: | Matho, M.H, Schlossman, A.M, Zajonc, D.M. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer.
Plos Pathog., 11, 2015
|
|
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | Descriptor: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0V
 
 | | Crystal Structure of Yeast Rtt107 | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.314 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0Y
 
 | | Crystal Structure of Yeast Rtt107 and Slx4 | | Descriptor: | Peptide from Structure-specific endonuclease subunit SLX4, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
6J0X
 
 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
4M1G
 
 | | Structure of murine IgG2a A27D7-Fab in complex with vaccinia antigen A33R at the resolution of 1.6 Angstroms | | Descriptor: | A33R, Murine IgG2a A27D7 Heavy chain Fab domain, Murine IgG2a A27D7 Light chain Fab domain, ... | | Authors: | Matho, M.H, Schlossman, A.M, Zajonc, D.M. | | Deposit date: | 2013-08-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer.
Plos Pathog., 11, 2015
|
|
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5USL
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv304 | | Descriptor: | Fab vv304 Heavy chain, Fab vv304 Light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
2KOD
 
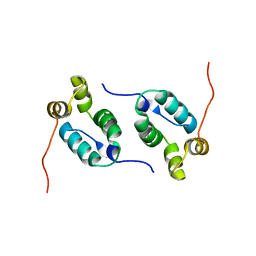 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
4EEE
 
 | |
4E9O
 
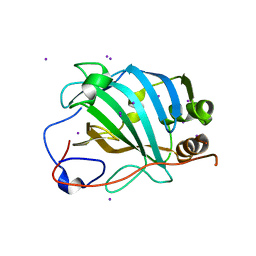 | | Vaccinia D8L ectodomain structure | | Descriptor: | IMV membrane protein, IODIDE ION | | Authors: | Matho, M.H, Zajonc, D.M. | | Deposit date: | 2012-03-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and Biochemical Characterization of the Vaccinia Virus Envelope Protein D8 and Its Recognition by the Antibody LA5.
J.Virol., 86, 2012
|
|
4EBQ
 
 | |
4EKX
 
 | |
4ETQ
 
 | |
8HGH
 
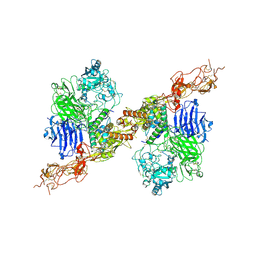 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
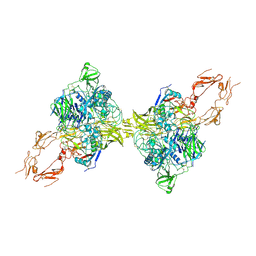 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HR2
 
 | | Ternary Crystal Complex Structure of RBD with NB1B5 and NB1C6 | | Descriptor: | NB1B5, NB1C6, Spike protein S1 | | Authors: | Sun, Z. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure basis of two nanobodies neutralizing SARS-CoV-2 Omicron variant by targeting ultra-conservative epitopes.
J.Struct.Biol., 215, 2023
|
|
7Y5N
 
 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
