2OPW
 
 | | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo) | | Descriptor: | PHYHD1 protein | | Authors: | Zhang, Z, Butler, D, McDonough, M.A, Kavanagh, K.L, Bray, J.E, Ng, S.S, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human phytanoyl-CoA dioxygenase PHYHD1 (apo)
To be Published
|
|
2OQ6
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2R37
 
 | | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant) | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 3, SODIUM ION | | Authors: | Pilka, E.S, Guo, K, Gileadi, O, Rojkowa, A, von Delft, F, Pike, A.C.W, Kavanagh, K.L, Johannson, C, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-29 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human glutathione peroxidase 3 (selenocysteine to glycine mutant).
TO BE PUBLISHED
|
|
2R4H
 
 | | Crystal structure of a C1190S mutant of the 6th PDZ domain of human membrane associated guanylate kinase | | Descriptor: | HISTIDINE, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ugochukwu, E, Pilka, E.S, Hozjan, V, Kavanagh, K.L, Cooper, C, Pike, A.C.W, Elkins, J.M, Doyle, D.A, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-31 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a C1190S mutant of the 6th PDZ domain of human membrane associated guanylate kinase.
To be Published
|
|
2C2N
 
 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
2C43
 
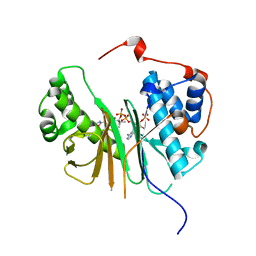 | | STRUCTURE OF AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE- PHOSPHOPANTETHEINYL TRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Oppermann, U. | | Deposit date: | 2005-10-14 | | Release date: | 2005-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
1YDE
 
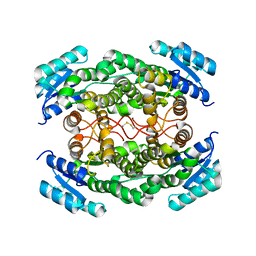 | | Crystal Structure of Human Retinal Short-Chain Dehydrogenase/Reductase 3 | | Descriptor: | Retinal dehydrogenase/reductase 3 | | Authors: | Lukacik, P, Bunkozci, G, Kavanagh, K, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity.
Biochem.J., 402, 2007
|
|
4KFA
 
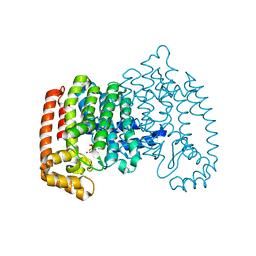 | | Crystal structure of human farnesyl pyrophosphate synthase (t201a mutant) complexed with mg and zoledronate | | Descriptor: | 1,2-ETHANEDIOL, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C, Walter, R.L. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of human farnesyl pyrophosphate synthase (t201a mutant) complexed with mg and zoledronate
To be Published
|
|
4KQ5
 
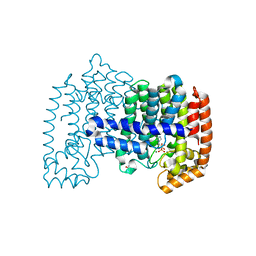 | |
4KQS
 
 | | Crystal Structure of Farnesyl Pyrophosphate Synthase Mutant (Y204A) Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Farnesyl Pyrophosphate Synthase Mutant (Y204A) Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate
To be Published
|
|
6Q94
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
3PPG
 
 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, alanine variant with zinc | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, ZINC ION | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
3PPC
 
 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, tyrosine variant with zinc | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, CHLORIDE ION, ZINC ION | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
3PPF
 
 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, alanine variant without zinc | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
3PPH
 
 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, threonine variant | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
3QNF
 
 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2JFD
 
 | | Structure of the MAT domain of human FAS | | Descriptor: | FATTY ACID SYNTHASE | | Authors: | Bunkoczi, G, Kavanagh, K, Hozjan, V, Rojkova, A, Wu, X, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Smith, S, Oppermann, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for different specificities of acyltransferases associated with the human cytosolic and mitochondrial fatty acid synthases.
Chem. Biol., 16, 2009
|
|
4CCL
 
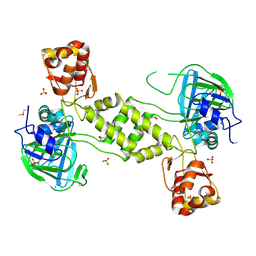 | | X-Ray structure of E. coli ycfD | | Descriptor: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | Authors: | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4BXF
 
 | |
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCN
 
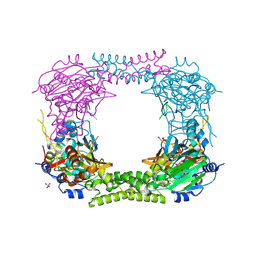 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 L299C/C300S) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-2) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCM
 
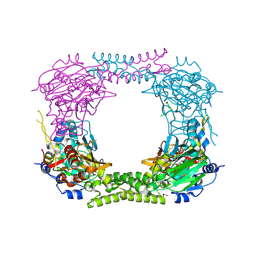 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCO
 
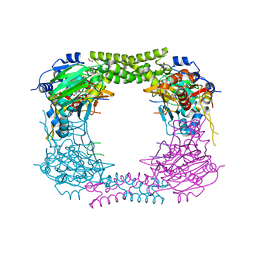 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 S373C) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G214C) peptide fragment (complex-3) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
