8R1T
 
 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
8R1P
 
 | |
8R1N
 
 | |
8R18
 
 | | Pim1 in complex with (E)-4-(4-hydroxystyryl)benzoic acid and Pimtide | | Descriptor: | (E)-4-(4-hydroxystyryl)benzoic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
8R10
 
 | |
8BJL
 
 | |
2NSO
 
 | | Trna-gunanine-transglycosylase (TGT) mutant Y106F, C158V, A232S, V233G- APO-Structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-06 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor.
Plos One, 8, 2013
|
|
2NVD
 
 | | Human Aldose Reductase complexed with novel naphtho[1,2-d]isothiazole acetic acid derivative (2) | | Descriptor: | 2-(CARBOXYMETHYL)-1-OXO-1,2-DIHYDRONAPHTHO[1,2-D]ISOTHIAZOLE-4-CARBOXYLIC ACID 3,3-DIOXIDE, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-11-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evidence for a novel binding site conformer of aldose reductase in ligand-bound state
J.Mol.Biol., 369, 2007
|
|
2NVC
 
 | | Human Aldose Reductase complexed with novel naphtho[1,2-d]isothiazole acetic acid derivative (3) | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {4-[(CARBOXYMETHOXY)CARBONYL]-3,3-DIOXIDO-1-OXONAPHTHO[1,2-D]ISOTHIAZOL-2(1H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-11-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence for a novel binding site conformer of aldose reductase in ligand-bound state
J.Mol.Biol., 369, 2007
|
|
4YXO
 
 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (3). | | Descriptor: | 4-ethylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
4YYT
 
 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (5). | | Descriptor: | 4-(2-hydroxyethyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
4YXI
 
 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (2). | | Descriptor: | 4-methylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
4YXU
 
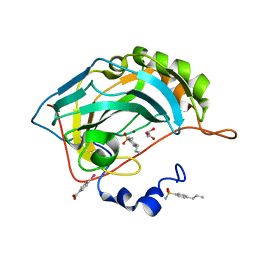 | | Human Carbonic Anhydrase II complexed with an inhibitor with a benzenesulfonamide group (4). | | Descriptor: | 4-propylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Kinetic and Structural Insights into the Mechanism of Binding of Sulfonamides to Human Carbonic Anhydrase by Computational and Experimental Studies.
J.Med.Chem., 59, 2016
|
|
6FPU
 
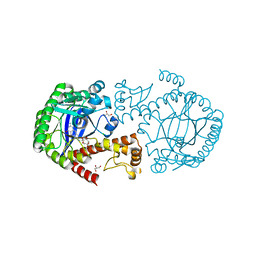 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-((((3aS,5aR,8bS)-2,2,7,7-tetramethyltetrahydro-3aH-bis([1,3]dioxolo)[4,5-b:4',5'-d]pyran-3a-yl)methyl)amino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-azanyl-2-[[(1~{R},2~{S},6~{S},9~{R})-4,4,11,11-tetramethyl-3,5,7,10,12-pentaoxatricyclo[7.3.0.0^{2,6}]dodecan-6-yl]methylamino]-3,7-dihydroimidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2018-02-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Sugar Acetonides are a Superior Motif for Addressing the Large, Solvent-Exposed Ribose-33 Pocket of tRNA-Guanine Transglycosylase.
Chemistry, 24, 2018
|
|
1OQ5
 
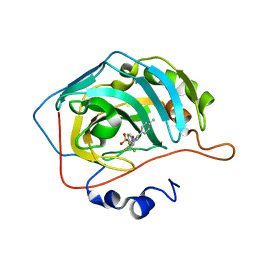 | | CARBONIC ANHYDRASE II IN COMPLEX WITH NANOMOLAR INHIBITOR | | Descriptor: | 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, Carbonic anhydrase II, ZINC ION | | Authors: | Weber, A, Casini, A, Heine, A, Kuhn, D, Supuran, C.T, Scozzafava, A, Klebe, G. | | Deposit date: | 2003-03-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected nanomolar inhibition of carbonic anhydrase by COX-2-selective celecoxib: new pharmacological opportunities due to related binding site recognition.
J.Med.Chem., 47, 2004
|
|
4YS1
 
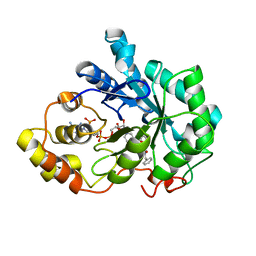 | | Human Aldose Reductase complexed with a ligand with an IDD structure (2) at 1.07 A. | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Price for Opening the Transient Specificity Pocket in Human Aldose Reductase upon Ligand Binding: Structural, Thermodynamic, Kinetic, and Computational Analysis.
ACS Chem. Biol., 12, 2017
|
|
8AFR
 
 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
3FLF
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, GLYCEROL, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-valyl-L-leucine, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2008-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: binding of phosphonamidate to the S1'-pocket of thermolysin.
Biochim.Biophys.Acta, 1800, 2010
|
|
3BIU
 
 | | Human thrombin-in complex with UB-THR10 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
5EGR
 
 | | tRNA guanine transglycosylase (TGT) in complex with an Immucillin derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-7-[(2~{S},3~{R},5~{S})-5-(hydroxymethyl)-3-oxidanyl-pyrrolidin-2-yl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of an Immucillin Derivative as a Class of tRNA-Guanine Transglycosylase Inhibitors: Exploration of a Transition State Analogous Binding Mode
To be Published
|
|
3FV4
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Displacement of disordered water molecules from hydrophobic pocket creates enthalpic signature: binding of phosphonamidate to the S1'-pocket of thermolysin.
Biochim.Biophys.Acta, 1800, 2010
|
|
3FVP
 
 | | Thermolysin inhibition | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Englert, L, Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2009-01-16 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Thermolysin inhibition
To be Published
|
|
3BIV
 
 | | Human thrombin-in complex with UB-THR11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
8AUU
 
 | |
8AQG
 
 | |
