9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
1VCQ
 
 | |
8GK7
 
 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.
Nat Microbiol, 9, 2024
|
|
8H08
 
 | |
8H07
 
 | |
1VCP
 
 | |
7X2H
 
 | |
7XD2
 
 | |
5GMP
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with XTF-262 | | Descriptor: | Epidermal growth factor receptor, N-[3-[2-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-5-methyl-7-oxidanylidene-pyrido[2,3-d]pyrimidin-8-yl]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | A structure-guided optimization of pyrido[2,3-d]pyrimidin-7-ones as selective inhibitors of EGFR(L858R/T790M) mutant with improved pharmacokinetic properties.
Eur J Med Chem, 126, 2017
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
6JRK
 
 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRJ
 
 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
2YGW
 
 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
3BK9
 
 | | H55A mutant of tryptophan 2,3-dioxygenase from Xanthomonas campestris | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Bruckmann, C, Mowat, C.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
4TUW
 
 | |
4TV0
 
 | |
1COV
 
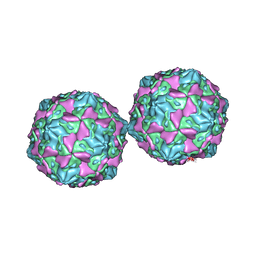 | | COXSACKIEVIRUS B3 COAT PROTEIN | | Descriptor: | COXSACKIEVIRUS COAT PROTEIN, MYRISTIC ACID, PALMITIC ACID | | Authors: | Muckelbauer, J.K, Rossmann, M.G. | | Deposit date: | 1994-10-19 | | Release date: | 1996-03-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure determination of coxsackievirus B3 to 3.5 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
1EP9
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CONFORMATIONAL CHANGE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
1FVO
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE TRANSCARBAMYLASE COMPLEXED WITH CARBAMOYL PHOSPHATE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-09-20 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
7VRA
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | Descriptor: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7VRE
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | Descriptor: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
2KFP
 
 | | Solution NMR structure of PSPTO_3016 from Pseudomonas syringae. Northeast Structural Genomics Consortium target PsR293. | | Descriptor: | PSPTO_3016 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Zhao, L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
4NOO
 
 | | Molecular mechanism for self-protection against type VI secretion system in Vibrio cholerae | | Descriptor: | Putative uncharacterized protein, VgrG protein | | Authors: | Yang, X, Xu, M, Jiang, T, Fan, Z. | | Deposit date: | 2013-11-20 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism for self-protection against the type VI secretion system in Vibrio cholerae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1MOL
 
 | |
