4P5K
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergy and Autoimmunity | | Descriptor: | HLA class II histocompatibility antigen, DP alpha 1 chain, RAS peptide,HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Wang, Y, Crawford, F, Novikov, A, Wimberly, B.T, Kieft, J.S, Falta, M.T, Bowerman, N.A, Marrack, P, Fontenot, A.P, Dai, S, Kappler, J.W. | | Deposit date: | 2014-03-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4EYR
 
 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
4KO5
 
 | | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F) | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Inampudi, K.K, Wang, Y, Meng, W, Tobin, P.H, Wilson, C.J. | | Deposit date: | 2013-05-11 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F)
To be Published
|
|
4GYE
 
 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | Descriptor: | P1F peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
8H0O
 
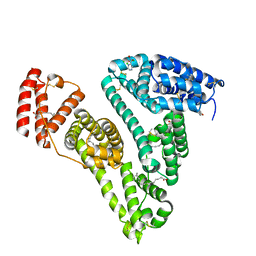 | | Crystal structure of human serum albumin and ruthenium PZA complex adduct | | Descriptor: | Albumin, CHLORIDE ION, NITRIC OXIDE, ... | | Authors: | Gong, W.J, Wang, Y, Bai, H.H, Wang, W.M, Wang, H.F. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium PZA complex adduct
To Be Published
|
|
4EKU
 
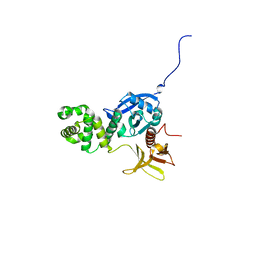 | |
4GZF
 
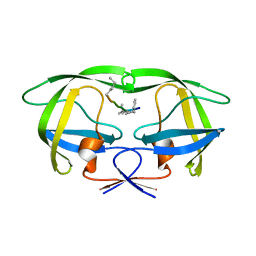 | | Multi-drug resistant HIV-1 protease 769 variant with reduced LrF peptide | | Descriptor: | LrF peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Kovari, I.A, Brunzelle, J.S, Reiter, S.J, Kovari, L.C. | | Deposit date: | 2012-09-06 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
4L1A
 
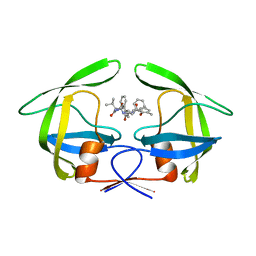 | | Crystallographic study of multi-drug resistant HIV-1 protease Lopinavir complex: mechanism of drug recognition and resistance | | Descriptor: | MDR769 HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Dewdney, T, Reiter, S, Brunzelle, J, Kovari, I, Kovari, L. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of multi-drug resistant HIV-1 protease lopinavir complex: mechanism of drug recognition and resistance.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
4LOF
 
 | | Human p53 Core Domain Mutant V157F/N235K/N239Y | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LOE
 
 | | Human p53 Core Domain Mutant N239Y | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4EG2
 
 | | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine | | Descriptor: | ACETATE ION, Cytidine deaminase, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine.
TO BE PUBLISHED
|
|
4KVP
 
 | | Human p53 Core Domain Mutant V157F | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
4LO9
 
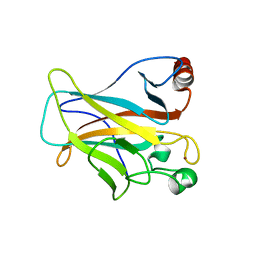 | | Human p53 Core Domain Mutant N235K | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5XG7
 
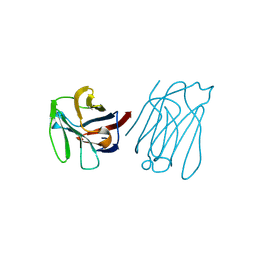 | | Galectin-13/Placental Protein 13 crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y, Wang, Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
8YM2
 
 | | Crystal structure of AIDA-1 PTB domain in complex with SynGAP NPxF motif | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B, Ras/Rap GTPase-activating protein SynGAP | | Authors: | Wang, X, Wang, Y, Cai, Q, Zhang, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AIDA-1/ANKS1B Binds to the SynGAP Family RasGAPs with High Affinity and Specificity.
J.Mol.Biol., 436, 2024
|
|
7E35
 
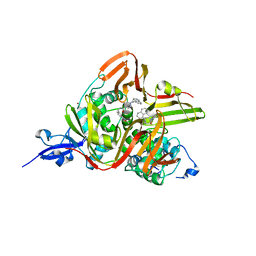 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
5XHC
 
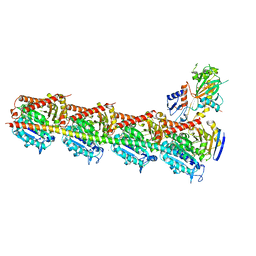 | | Crystal structure of T2R-TTL-PO10 complex | | Descriptor: | (3Z,6Z)-3-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]-6-[[3-(4-fluorophenyl)carbonylphenyl]methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Chu, Y, Wang, Y, Yang, J, Li, W. | | Deposit date: | 2017-04-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
5XI5
 
 | | Crystal structure of T2R-TTL-PO5 complex | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Chu, Y, Wang, Y, Yang, J, Li, W. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
7F0X
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F12
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F15
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
8WQR
 
 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6K3Z
 
 | | Crystal structure of dCas9 in complex with sgRNA and DNA (TGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (28-MER), DNA (5'-D(*AP*AP*AP*TP*GP*AP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-22 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
