4CB4
 
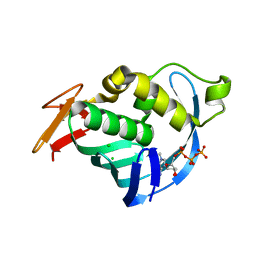 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
1JKM
 
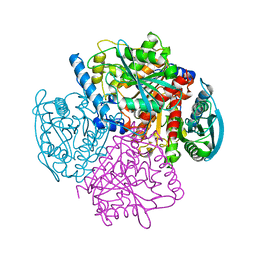 | | BREFELDIN A ESTERASE, A BACTERIAL HOMOLOGUE OF HUMAN HORMONE SENSITIVE LIPASE | | Descriptor: | BREFELDIN A ESTERASE | | Authors: | Wei, Y, Contreras, A.J, Sheffield, P, Osterlund, T, Derewenda, U, Matern, U.O, Derewenda, Z.S. | | Deposit date: | 1998-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase.
Nat.Struct.Biol., 6, 1999
|
|
4A2W
 
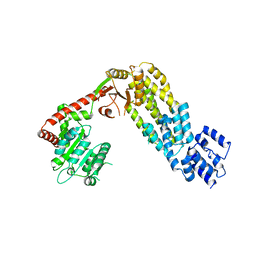 | | Structure of full-length duck RIG-I | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A36
 
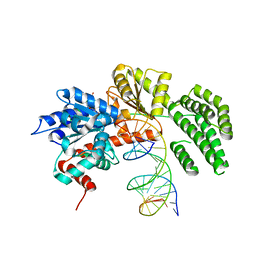 | | Structure of duck RIG-I helicase domain bound to 19-mer dsRNA and ATP transition state analogue | | Descriptor: | 5'-R(*GP*CP*AP*UP*GP*CP*GP*AP*CP*CP*UP*CP*UP*GP *UP*UP*UP*GP*A)-3', 5'-R(*UP*CP*AP*AP*AP*CP*AP*GP*AP*GP*GP*UP*CP*GP *CP*AP*UP*GP*C)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2V
 
 | | Structure of duck RIG-I C-terminal domain (CTD) | | Descriptor: | GLYCEROL, RETINOIC ACID INDUCIBLE PROTEIN I, ZINC ION | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2P
 
 | | Structure of duck RIG-I helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
4A2X
 
 | | Structure of duck RIG-I C-terminal domain (CTD) with 14-mer dSRNA | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*UP*GP*UP*UP*CP*UP*CP*CP*CP)-3', 5'-R(*GP*GP*GP*AP*GP*AP*AP*CP*AP*AP*CP*GP*CP*GP)-3', RETINOIC ACID INDUCIBLE PROTEIN I, ... | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA.
Cell, 147, 2011
|
|
4A2Q
 
 | | Structure of duck RIG-I tandem CARDs and helicase domain | | Descriptor: | RETINOIC ACID INDUCIBLE PROTEIN I | | Authors: | Kowalinski, E, Lunardi, T, McCarthy, A.A, Cusack, S. | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for the Activation of Innate Immune Pattern Recognition Receptor Rig-I by Viral RNA.
Cell(Cambridge,Mass.), 147, 2011
|
|
6RTD
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6TPO
 
 | | Conformation of cd1 nitrite reductase NirS without bound heme d1 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, HEME C, Nitrite reductase, ... | | Authors: | Kluenemann, T, Blankenfeldt, W. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structure of heme d 1 -free cd 1 nitrite reductase NirS.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TP9
 
 | | c-type cytochrome NirC | | Descriptor: | Cytochrome c55X, HEME C | | Authors: | Kluenemann, T, Henke, S, Blankenfeldt, W. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TSI
 
 | |
6TV9
 
 | |
6TV2
 
 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
4GCR
 
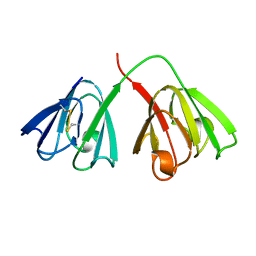 | | STRUCTURE OF THE BOVINE EYE LENS PROTEIN GAMMA-B (GAMMA-II)-CRYSTALLIN AT 1.47 ANGSTROMS | | Descriptor: | GAMMA-B CRYSTALLIN | | Authors: | Slingsby, C, Najmudin, S, Nalini, V, Driessen, H.P.C, Blundell, T.L, Moss, D.S, Lindley, P. | | Deposit date: | 1992-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the bovine eye lens protein gammaB(gammaII)-crystallin at 1.47 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7MY2
 
 | |
7MY3
 
 | |
7Z7C
 
 | |
7PZ1
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH8535 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-bromanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(3-methoxy-4-methyl-phenyl)piperidine-1-carboxamide, GLYCEROL, ... | | Authors: | Scaletti, E.R, Helleday, T, Stenmark, P. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
2XSQ
 
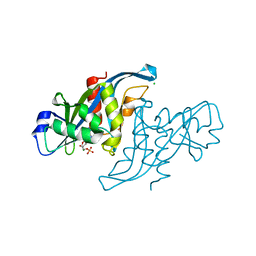 | | Crystal structure of human Nudix motif 16 (NUDT16) in complex with IMP and magnesium | | Descriptor: | CHLORIDE ION, INOSINIC ACID, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van den Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P. | | Deposit date: | 2010-09-29 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Specificity of Human Nudt16 and its Regulation by Inosine Monophosphate.
Plos One, 10, 2015
|
|
8I3G
 
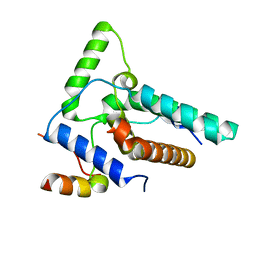 | | Crystal structure of Eaf3-Eaf7 complex | | Descriptor: | Chromatin modification-related protein EAF3, Chromatin modification-related protein EAF7 | | Authors: | Chen, Z, Xu, C. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for Eaf3-mediated assembly of Rpd3S and NuA4.
Cell Discov, 9, 2023
|
|
8I3F
 
 | |
6G40
 
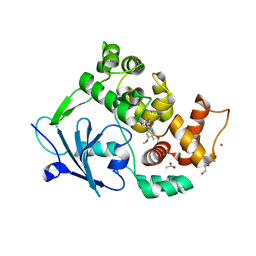 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH9525 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Optimization of N-Piperidinyl-Benzimidazolone Derivatives as Potent and Selective Inhibitors of 8-Oxo-Guanine DNA Glycosylase 1.
Chemmedchem, 18, 2023
|
|
5I7R
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 2 [3-(3-([1,1'-biphenyl]-3-yl)ureido)benzoic acid] | | Descriptor: | 3-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}benzoic acid, ACETATE ION, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
