7Z13
 
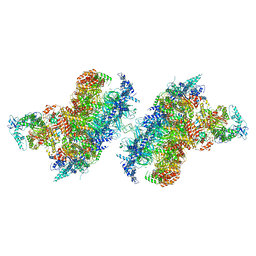 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
7QHS
 
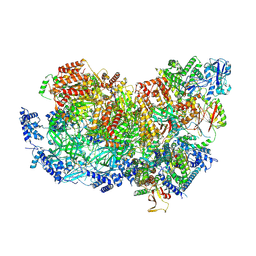 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
8FEN
 
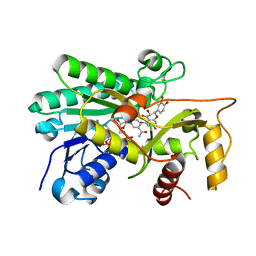 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEM
 
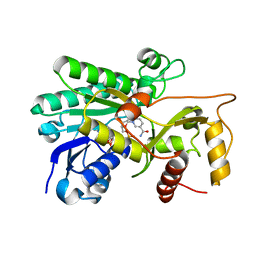 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
1BE9
 
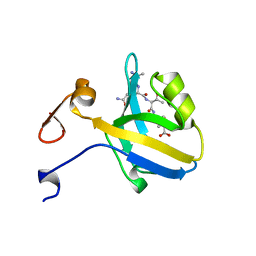 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BFE
 
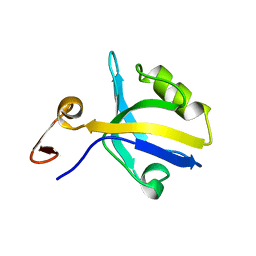 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
5T88
 
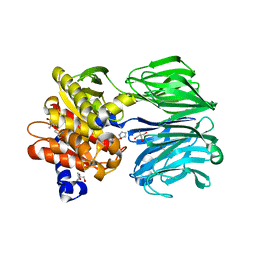 | | Prolyl oligopeptidase from Pyrococcus furiosus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, PROLINE, ... | | Authors: | Ellis-Guardiola, K, Lewis, J, Sukumar, N. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure and Conformational Dynamics of Pyrococcus furiosus Prolyl Oligopeptidase.
Biochemistry, 58, 2019
|
|
5V30
 
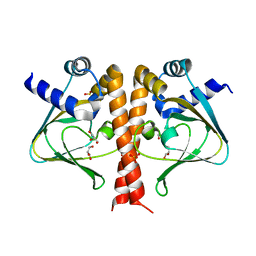 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NP6
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6P2V
 
 | | RebH Variant 10S, Tryptamine 5-halogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase RebH | | Authors: | Andorfer, M.C, Sukumar, N, Lewis, J.C. | | Deposit date: | 2019-05-22 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural and Computational Analysis of Laboratory-Evolved Halogenases Reveals Molecular Details of Site-Selectivity
To Be Published
|
|
6P00
 
 | | RebH Variant 8F, Tryptamine 6-halogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase RebH | | Authors: | Andorfer, M.C, Sukumar, N, Lewis, J.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural and Computational Analysis of Laboratory-Evolved Halogenases Reveals Molecular Details of Site-Selectivity
To Be Published
|
|
6CAN
 
 | |
2LZG
 
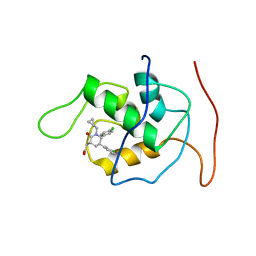 | | NMR Structure of Mdm2 (6-125) with Pip-1 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-(cyclopropylmethyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Michelsen, K.B, Jordan, J.B, Lewis, J, Long, A.M, Yang, E, Rew, Y, Zhou, J, Yakowec, P, Schnier, P.D, Huang, X, Poppe, L. | | Deposit date: | 2012-10-02 | | Release date: | 2012-11-07 | | Method: | SOLUTION NMR | | Cite: | Ordering of the N-Terminus of Human MDM2 by Small Molecule Inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
7JLY
 
 | |
3T22
 
 | |
7JU0
 
 | |
2AGN
 
 | | Fitting of hepatitis C virus internal ribosome entry site domains into the 15 A Cryo-EM map of a HCV IRES-80S ribosome (H. sapiens) complex | | Descriptor: | 6 nt A-RNA helix, HCV IRES DOMAIN II, HCV IRES IIIABC, ... | | Authors: | Boehringer, D, Thermann, R, Ostareck-Lederer, A, Lewis, J.D, Stark, H. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES
Structure, 13, 2005
|
|
3HO7
 
 | |
4CB6
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 11) | | Descriptor: | 2,9-N,N-DI(4-CARBOXYBUTYL)-7-N-METHYLGUANINE, IODIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
2VQZ
 
 | | Structure of the cap-binding domain of influenza virus polymerase subunit PB2 with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Tarendeau, F, Resa-Infante, P, Coloma, R, Crepin, T, Sehr, P, Lewis, J, Ruigrok, R.W.H, Ortin, J, Hart, D.J, Cusack, S. | | Deposit date: | 2008-03-21 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for CAP Binding by Influenza Virus Polymerase Subunit Pb2.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4CB4
 
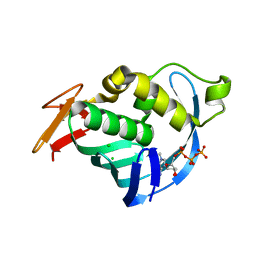 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB5
 
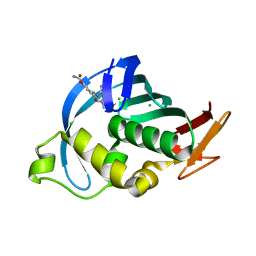 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8f) | | Descriptor: | 9-N-(3-CARBOXY-4-HYDROXYPHENYL)KETOMETHYL-7-N-METHYLGUANINE, CHLORIDE ION, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CB7
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8e) | | Descriptor: | 5-[2-(2-azanyl-7-methyl-6-oxidanylidene-1H-purin-9-ium-9-yl)ethanoyl]-2-oxidanyl-benzoic acid, GLYCEROL, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
2MHS
 
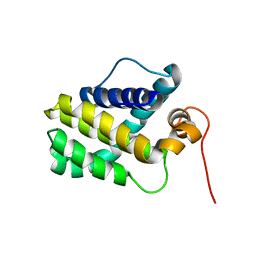 | | NMR Structure of human Mcl-1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Liu, G, Poppe, L, Aoki, K, Yamane, H, Lewis, J, Szyperski, T. | | Deposit date: | 2013-12-04 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High-Quality NMR Structure of Human Anti-Apoptotic Protein Domain Mcl-1(171-327) for Cancer Drug Design.
Plos One, 9, 2014
|
|
3UKI
 
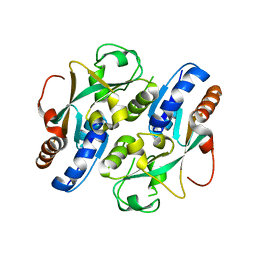 | | Crystal structure of reduced OxyR from Porphyromonas gingivalis | | Descriptor: | OxyR | | Authors: | Svintradze, D.V, Wright, H.T, Collazo-Santiago, E.A, Lewis, J.P. | | Deposit date: | 2011-11-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structures of the Porphyromonas gingivalis OxyR regulatory domain explain differences in expression of the OxyR regulon in Escherichia coli and P. gingivalis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
