7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSJ
 
 | | Crystal structure of 1,6-APD (2-ANIMOPHENOL-1,6-DIOXYGENASE) complexed with intermediate products | | Descriptor: | (2Z,4Z)-2-imino-6-oxohex-4-enoic acid, (3E)-3-iminooxepin-2(3H)-one, 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSH
 
 | | Crystal structure of native 1,6-APD (with Iron), 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, FE (II) ION | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8G3C
 
 | |
8G3E
 
 | | Crystal structure of human WDR5 in complex with (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide (compound 2, WDR5-MYC inhibitor) | | Descriptor: | (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Zhao, M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery and Structure-Based Design of Inhibitors of the WD Repeat-Containing Protein 5 (WDR5)-MYC Interaction.
J.Med.Chem., 66, 2023
|
|
2FX4
 
 | |
7L5B
 
 | |
7KS9
 
 | |
2FX6
 
 | | bovine trypsin complexed with 2-aminobenzamidazole | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, Trypsin | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FWW
 
 | |
2FPZ
 
 | | Human tryptase with 2-amino benzimidazole | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FS8
 
 | | Human beta-tryptase II with inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2G8A
 
 | | Lactobacillus casei Y261M in complex with substrate, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, thymidylate synthase | | Authors: | Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
Biochemistry, 45, 2006
|
|
2FS9
 
 | | Human beta tryptase II with inhibitor CRA-28427 | | Descriptor: | ETHYL {(1S)-5-AMINO-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]PENTYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FXR
 
 | | human beta tryptase II complexed with activated ketone inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-02-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
8GZ9
 
 | | Cryo-EM structure of Abeta2 fibril polymorph2 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8GZ8
 
 | | Cryo-EM structure of Abeta2 fibril polymorph1 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
5TRF
 
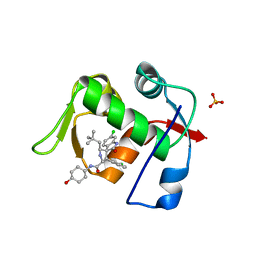 | | MDM2 in complex with SAR405838 | | Descriptor: | (2'S,3R,4'S,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(trans-4-hydroxycyclohexyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.
Cancer Res., 74, 2014
|
|
7LS9
 
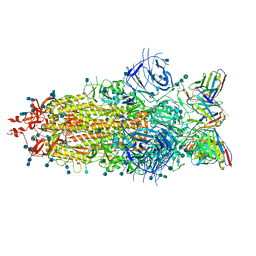 | |
7LSS
 
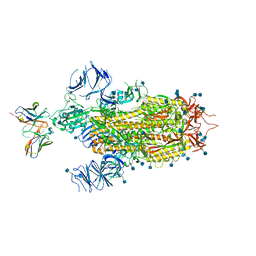 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-7 variable heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-17 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.
Structure, 29, 2021
|
|
3LKZ
 
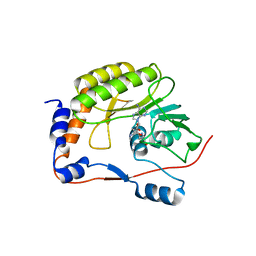 | |
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
