1SW0
 
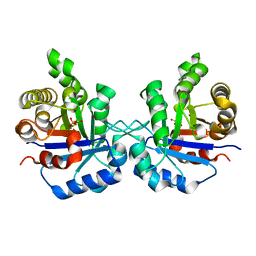 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1KS0
 
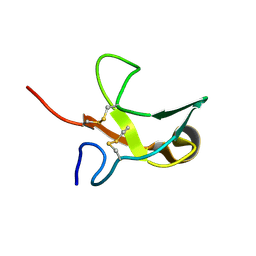 | | The First Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | Matrix Metalloproteinase 2 | | Authors: | Gehrmann, M, Briknarova, K, Banyai, L, Patthy, L, Llinas, M. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The col-1 module of human matrix metalloproteinase-2 (MMP-2): structural/functional relatedness between gelatin-binding fibronectin type II modules and lysine-binding kringle domains.
Biol.Chem., 383, 2002
|
|
3U10
 
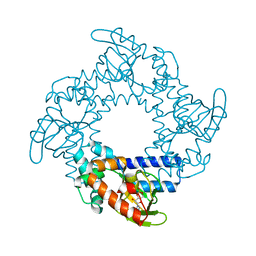 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
3U0Z
 
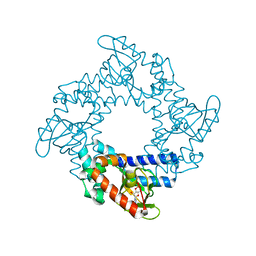 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4CML
 
 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CXL
 
 | | Human insulin analogue (D-ProB8)-insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
4CXN
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form I | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
138D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-HEXAGONAL CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
137D
 
 | | A-DNA DECAMER D(GCGGGCCCGC)-ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | DNA (5'-D(*GP*CP*GP*GP*GP*CP*CP*CP*GP*C)-3') | | Authors: | Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for crystal environment dominating base sequence effects on DNA conformation: crystal structures of the orthorhombic and hexagonal polymorphs of the A-DNA decamer d(GCGGGCCCGC) and comparison with their isomorphous crystal structures.
Biochemistry, 32, 1993
|
|
4CY7
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form II | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
1M6R
 
 | |
4GZK
 
 | |
4IQ6
 
 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
1MDG
 
 | | An Alternating Antiparallel Octaplex in an RNA Crystal Structure | | Descriptor: | 5'-R(*UP*(BGM)GP*AP*GP*GP*U)-3', COBALT HEXAMMINE(III), SODIUM ION | | Authors: | Pan, B.C, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction
Structure, 11, 2003
|
|
1JO2
 
 | |
2PLL
 
 | | Crystal structure of perdeuterated human arginase I | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, arginase-1 | | Authors: | Di Costanzo, L, Moulin, M, Haertlein, M, Meilleur, F, Christianson, D.W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification, assay, and crystal structure of perdeuterated human arginase I
Arch.Biochem.Biophys., 465, 2007
|
|
3TRK
 
 | | Structure of the Chikungunya virus nsP2 protease | | Descriptor: | Nonstructural polyprotein, SODIUM ION | | Authors: | Cheung, J, Franklin, M, Mancia, F, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure of the Chikungunya virus nsP2 protease
To be Published
|
|
1M77
 
 | | Near Atomic Resolution Crystal Structure of an A-DNA Decamer d(CCCGATCGGG): Cobalt Hexammine Interactions with A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*GP*AP*TP*CP*GP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Ramakrishnan, B, Sekharudu, C, Pan, B.C, Sundaralingam, M. | | Deposit date: | 2002-07-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Near-atomic resolution crystal structure of an A-DNA decamer d(CCCGATCGGG): cobalt hexammine interaction with A-DNA.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3QYU
 
 | | Crystal structure of human cyclophilin D at 1.54 A resolution at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F | | Authors: | Colliandre, L, Gelin, M, Labesse, G, Guichou, J.-F. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4I0C
 
 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
1FZQ
 
 | | CRYSTAL STRUCTURE OF MURINE ARL3-GDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 3, AMMONIUM ION, ... | | Authors: | Hillig, R.C, Hanzal-Bayer, M, Linari, M, Becker, J, Wittinghofer, A, Renault, L. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein.
Structure Fold.Des., 8, 2000
|
|
1JS2
 
 | | Crystal structure of C77S HiPIP: a serine ligated [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, high-potential iron protein | | Authors: | Mansy, S.S, Xiong, Y, Hemann, C, Hille, R, Sundaralingam, M, Cowan, J.A. | | Deposit date: | 2001-08-16 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and stability studies of C77S HiPIP: a serine ligated [4Fe-4S] cluster.
Biochemistry, 41, 2002
|
|
