8HVW
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
7DL2
 
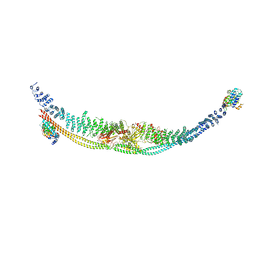 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
1MI1
 
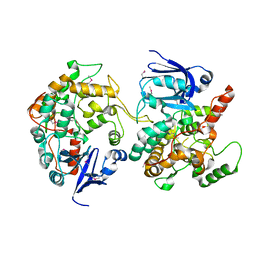 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
6LPF
 
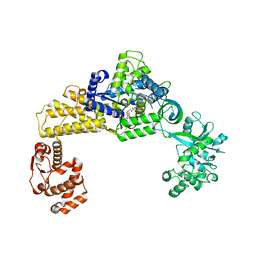 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
1TBU
 
 | |
6LR6
 
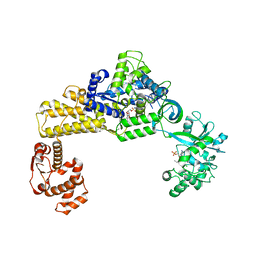 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6V4P
 
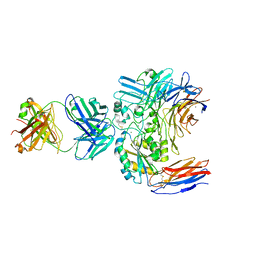 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
5V1D
 
 | |
1T77
 
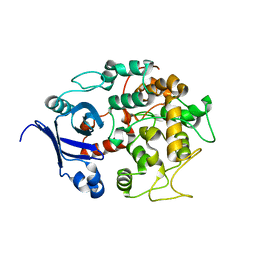 | | Crystal structure of the PH-BEACH domains of human LRBA/BGL | | Descriptor: | Lipopolysaccharide-responsive and beige-like anchor protein | | Authors: | Gebauer, D, Li, J, Jogl, G, Shen, Y, Myszka, D.G, Tong, L. | | Deposit date: | 2004-05-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the PH-BEACH Domains of Human LRBA/BGL
Biochemistry, 43, 2004
|
|
7DYS
 
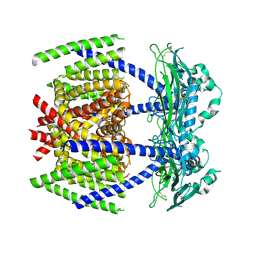 | |
8DSV
 
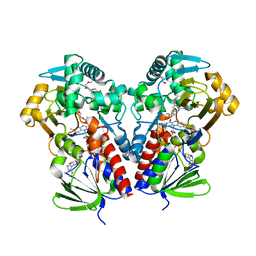 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
4LRH
 
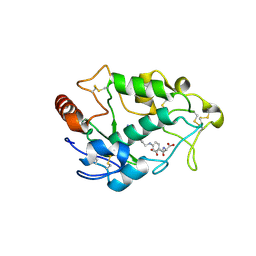 | | Crystal structure of human folate receptor alpha in complex with folic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, Folate receptor alpha | | Authors: | Ke, J, Chen, C, Zhou, X.E, Yi, W, Brunzelle, J.S, Li, J, Young, E.-L, Xu, H.E, Melcher, K. | | Deposit date: | 2013-07-19 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition of folic acid by folate receptors.
Nature, 500, 2013
|
|
8DQ8
 
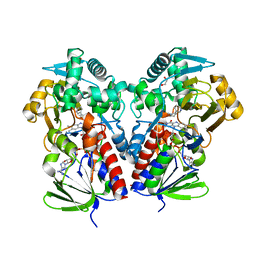 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
 | |
7RWR
 
 | | An RNA aptamer that decreases flavin redox potential | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (38-MER) | | Authors: | Gremminger, T, Li, J, Chen, S, Heng, X. | | Deposit date: | 2021-08-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA aptamer that shifts the reduction potential of metabolic cofactors.
Nat.Chem.Biol., 18, 2022
|
|
2LVN
 
 | | Structure of the gp78 CUE domain | | Descriptor: | E3 ubiquitin-protein ligase AMFR | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVO
 
 | | Structure of the gp78CUE domain bound to monubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
7CUN
 
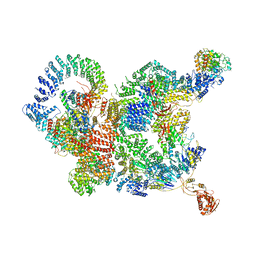 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
4NXF
 
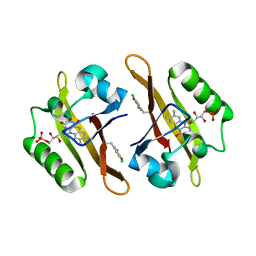 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
2LVP
 
 | | gp78CUE domain bound to the distal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2LVQ
 
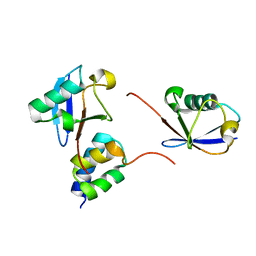 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
6KOE
 
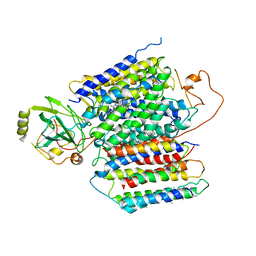 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOC
 
 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
