3THB
 
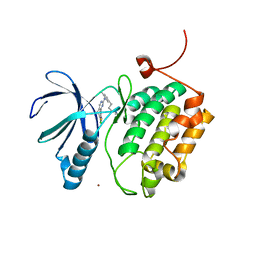 | | Structure of PLK1 kinase domain in complex with a benzolactam-derived inhibitor | | Descriptor: | 9-chloro-2-({5-[3-(dimethylamino)propyl]-2-methylpyridin-3-yl}amino)-5,7-dihydro-6H-pyrimido[5,4-d][1]benzazepine-6-thi one, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-08-18 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Benzolactam-Derived Inhibitor of Polo-Like Kinase 1 (MLN0905).
J.Med.Chem., 55, 2012
|
|
6FS2
 
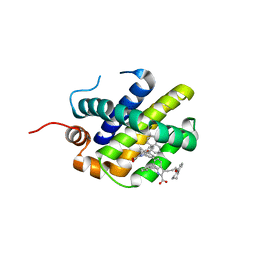 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS0
 
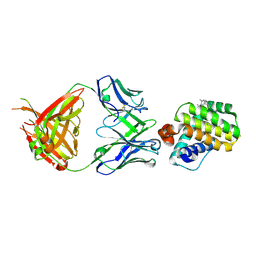 | |
5ECE
 
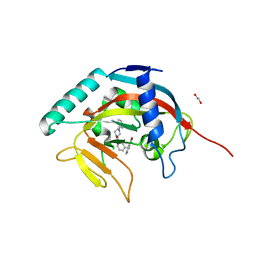 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3ZMM
 
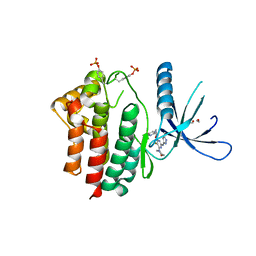 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5EBT
 
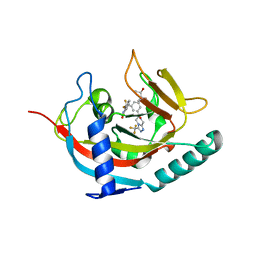 | | Tankyrase 1 with Phthalazinone 2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-[bis(fluoranyl)-[3-[[(6~{S})-6-methyl-3-(trifluoromethyl)-6,8-dihydro-5~{H}-[1,2,4]triazolo[4,3-a]pyrazin-7-yl]carbonyl]phenyl]methyl]-2~{H}-phthalazin-1-one, Tankyrase-1, ... | | Authors: | Kazmirski, S.L, Johannes, J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4GRB
 
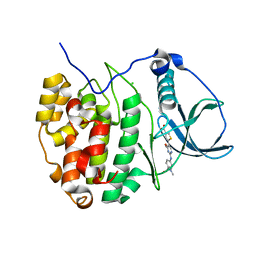 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
3U4U
 
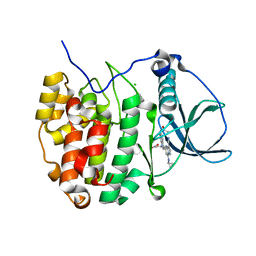 | | Casein kinase 2 in complex with AZ-Inhibitor | | Descriptor: | 3-{5-(acetylamino)-3-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]-1H-indol-1-yl}propanoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J. | | Deposit date: | 2011-10-10 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and Selective Inhibitors of CK2 Kinase Identified through Structure-Guided Hybridization.
ACS Med Chem Lett, 3, 2012
|
|
5XIW
 
 | | Crystal structure of T2R-TTL-Colchicine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-04-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
3IU3
 
 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
5I5D
 
 | | Salmonella global domain 245 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
5I5F
 
 | | Salmonella global domain 191 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
5I5H
 
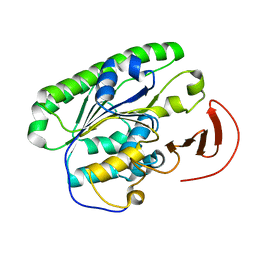 | | Ecoli global domain 245-586 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
2XA4
 
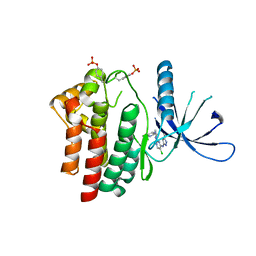 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-CHLORO-N2-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHYL]-N4-(5-METHYL-1H-PYRAZOL-3-YL)PYRIMIDINE-2,4-DIAMINE, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2010-03-26 | | Release date: | 2010-12-15 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of 5-Chloro-N2-[(1S)-1-(5-Fluoropyrimidin-2-Yl) Ethyl]-N4-(5-Methyl-1H-Pyrazol-3-Yl)Pyrimidine-2,4-Diamine (Azd1480) as a Novel Inhibitor of the Jak/Stat Pathway
J.Med.Chem., 54, 2011
|
|
4C61
 
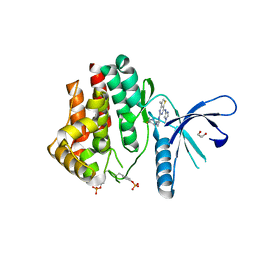 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | ACETATE ION, N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-7-methyl-N4-(1-methylimidazol-4-yl)thieno[3,2-d]pyrimidine-2,4-diamine, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J.A, Green, I, Pollard, H, Howard, T. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of 1-Methyl-1H-Imidazole Derivatives as Potent Jak2 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4C62
 
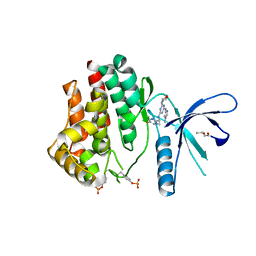 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | ACETATE ION, N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-n4-(1-methylimidazol-4-yl)-6-morpholino-1,3,5-triazine-2,4-diamine, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of 1-Methyl-1H-Imidazole Derivatives as Potent Jak2 Inhibitors.
J.Med.Chem., 57, 2014
|
|
5YL2
 
 | | Crystal structure of T2R-TTL-Y28 complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxy-3-oxidanyl-phenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Yang, T, Wen, J.L, Chen, L.J. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
7YI3
 
 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI2
 
 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI1
 
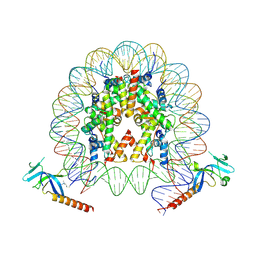 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
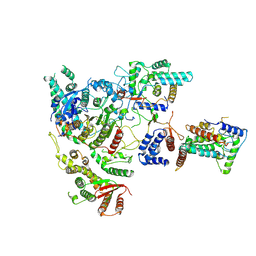 | | Cryo-EM structure of Rpd3S complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
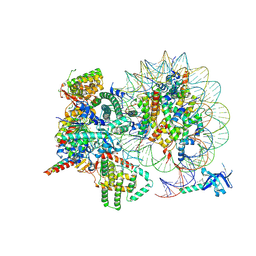 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI5
 
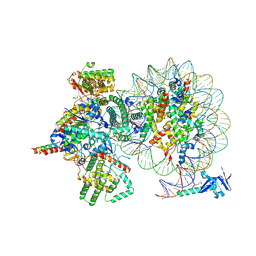 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7E74
 
 | |
7X88
 
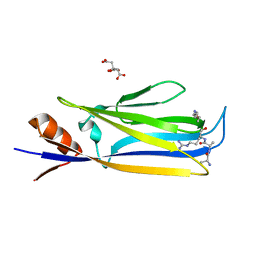 | |
